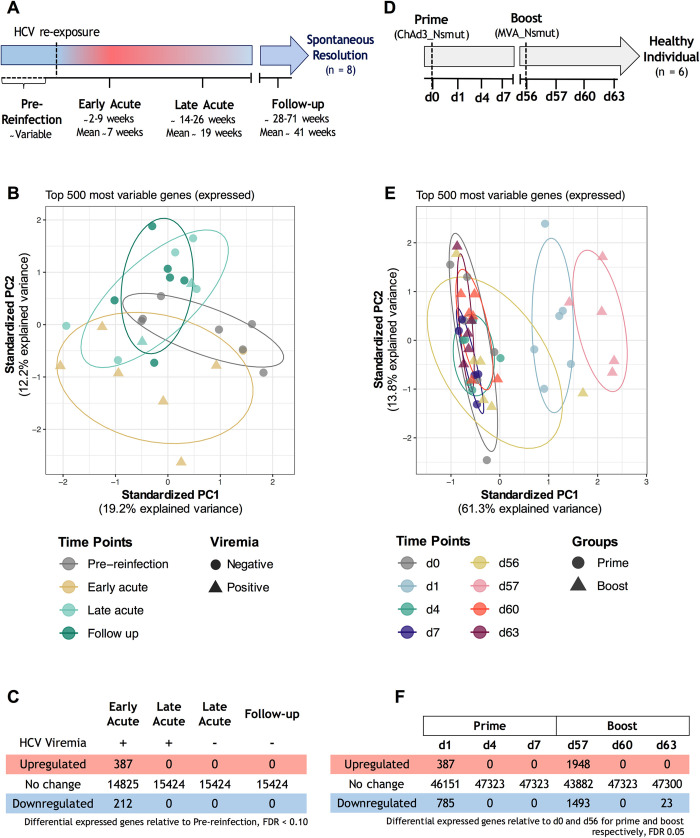
Fig 1. Study design and differential gene expression analysis.
(A) Reinfection study design. (B) Principal component analysis (PCA) of the top 500 genes expressed in the reinfection group. As described in Materials and Methods, patient-specific gene expression variation was removed using the function removeBatchEffect. Time points and groups are indicated by different colors and shapes, respectively. The ellipses denote the core area with a confidence interval of 68% for each group. (C) Number of differentially expressed genes (DEGs) across all reinfection time points. Pre-reinfection time-points were used as a baseline. Genes differentially expressed are selected using a Benjamin-Hochberg adjusted P value < 0.10. (D) Vaccine study design. Vaccine data were obtained from Hartnell et al. [18]. (E) PCA of the top 500 genes expressed in the vaccine group. (F) Number of DEGs across all vaccine samples. d0 and d56 were used as a baseline for prime and boost vaccine time points, respectively. Genes differentially expressed are selected using a Benjamin-Hochberg adjusted P value < 0.05.