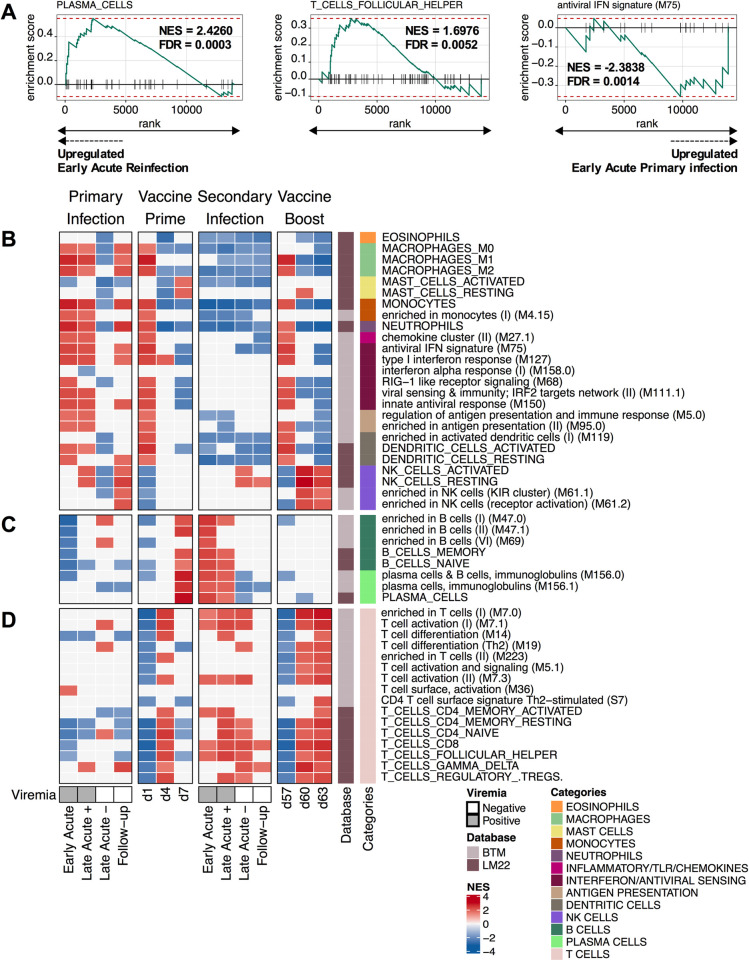
Fig 2. Upregulation of plasma cell modules at viremia positive time points during reinfection and their absence post-vaccine boosts.
(A) Enrichment plots depicting differential enrichment of LM22 (PLASMA_CELLS and T_CELLS_FOLLICULAR_HELPER) and BTM (antiviral IFN signature (M75)) modules for the Early acute time point of the reinfection ([R]) relative to primary infection ([P]) using the following contrast: (REARLY-ACUTE−RPRE-REINFECTION)–(PEARLY-ACUTE−PPRE-INFECTION). Primary infection data were obtained from (Rosenberg et al, 2018). Each gene forming the indicated module is represented by vertical black lines along the x-axis. The green line connects enrichment scores (ES) indicated by the y-axis and genes. Red-dashed lines indicate the maximum and minimum ES reached. False discovery rate (FDR) and normalized ES (NES) are presented with each enrichment plot. (B-D) Immune cell subsets (LM22 and BTM datasets indicated on sidebar) transcriptomic signature enrichment among the list of ranked differentially expressed genes at primary infection (Resolvers; n = 6), prime vaccine timepoints (n = 6), secondary infection (Resolvers; n = 7) and boost vaccine timepoints (n = 6). Pre-Infection times were used as a baseline for primary and secondary infections timepoints. d0 and d56 were used as a baseline for prime vaccine and boost vaccine time points, respectively. The x-axis indicates timepoints. The y-axis indicates immune subset enriched in at least one timepoint with a Benjamini−Hochberg-corrected gene set enrichment analysis (GSEA) p value < 0.05. Colors indicate the NES and show whether the immune subset was positively enriched (red) or negatively enriched (blue). Each module category is indicated on the color sidebar. Modules are further separated into three: (B) innate immune cells, (C) B cells and (D) T cells. The presence (Grey) or absence (White) of viral RNA is indicated below corresponding primary and secondary infection samples.