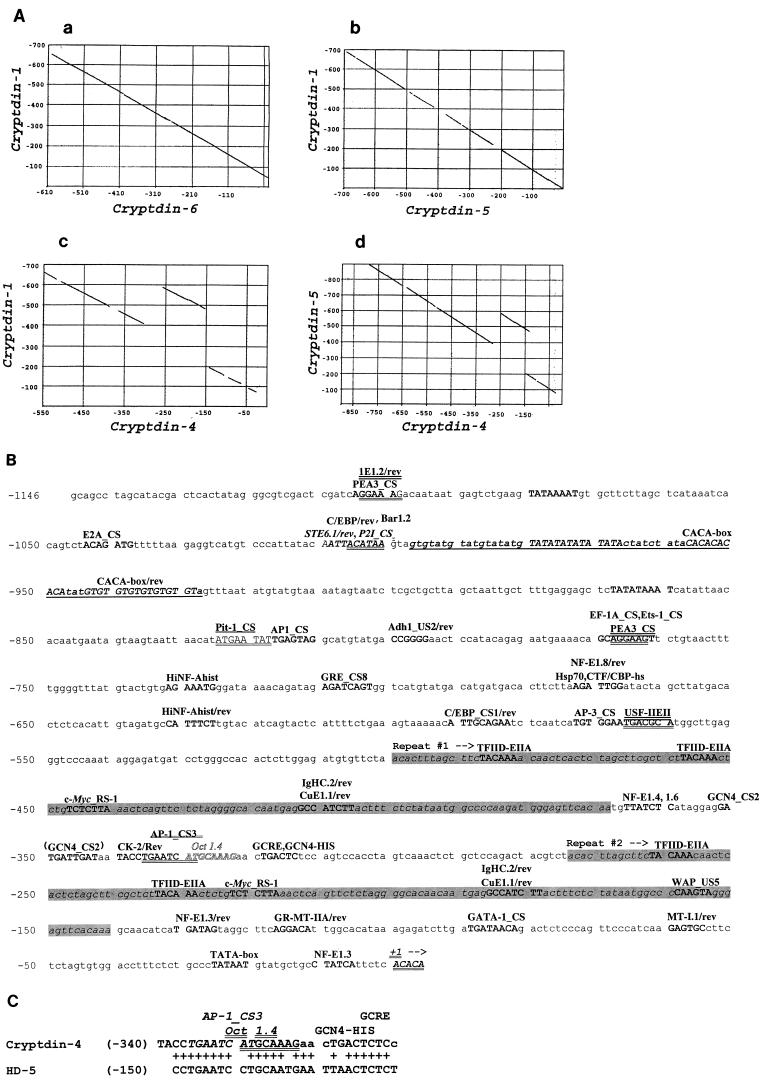
FIG. 5.
A repeated sequence element upstream of the cryptdin 4 gene transcriptional start site. (A) DNA sequences in the region from positions +1 to −700 upstream of the cryptdin genes shown were compared by the Pustell dot matrix alignment routine in the MacVector suite (see Materials and Methods). In contrast to comparisons of cryptdins 1 and 6 (a) as well as of cryptdins 1 and 5 (b), which were colinear and nearly identical, alignment comparisons involving the cryptdin 4 gene revealed a discontinuity in the sequence alignment that is indicative of a tandem repeat (c and d). (B) Potential transcription factor recognition sites (10, 11, 28) in the region upstream of the cryptdin 4 transcriptional start site are indicated in uppercase type and identified above lines of nucleotide sequence. The duplicated element described for panel A, from nucleotides −494 to −367 and −269 to −140, is indicated by shaded text. (C) The only region of similarity between the 1.2 kb of nucleotide sequence 5′ of the mouse cryptdin 4 and human HD-5 Paneth cell α-defensin gene transcription start sites is shown. Nucleotide sequences from the 5′ untranscribed regions of cryptdin 4 and HD-5 genes were aligned using the program Bestfit. Pluses between lines of DNA sequence denote identical nucleotide positions. Conserved potential DNA recognition sequences for AP-1 (italics), Oct 1.4 (double underlining), and GCRE/GCN4-HIS (single underlining) transcription factors are indicated.