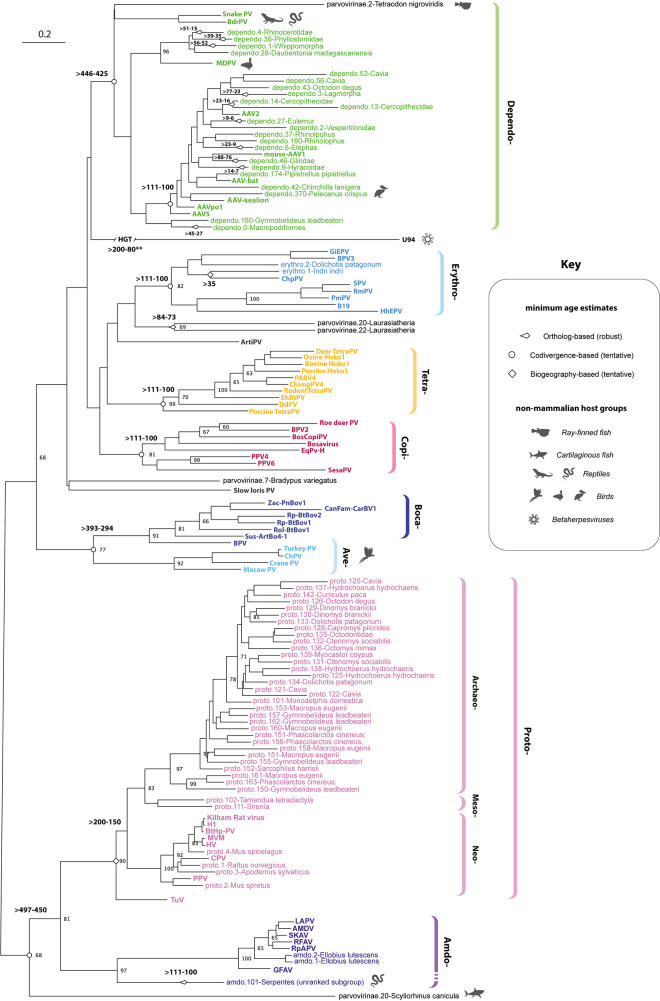
Fig 4. Evolution of subfamily Parvoviridae.
An ML phylogeny showing the reconstructed evolutionary relationships between contemporary parvoviruses of subfamily Parvovirinae and the EPVs derived from subfamily Parvovirinae. The phylogeny, which is midpoint rooted for display purposes, was reconstructed using an MSA spanning 270 amino acid residues in the Parvovirus Rep protein and the LG likelihood substitution model. Coloured brackets indicate the established parvovirus genera recognised by the International Committee for the Taxonomy of Viruses. Bootstrap support values (1,000 replicates) are shown for deeper internal nodes only. Scale bars show evolutionary distance in substitutions per site. Taxa labels are coloured based on taxonomic grouping as indicated by brackets; unclassified taxa are shown in black. Viral taxa are shown in bold, while EPV taxa are show in regular text. Numbers adjacent node shapes show minimum age estimates associated with lineages in millions of years before present (see Table 3). Abbreviations: AAV, adeno-associated virus; AMDV, Aleutian mink disease; BPV, bovine parvovirus; BrdPV, bearded dragon parvovirus; CPV, canine parvovirus; EPV, endogenous parvoviral element; HGT, horizontal gene transfer; HHV, human herpesvirus; MdPV, Muscovy duck parvovirus; ML, maximum likelihood; MSA, multiple sequence alignment; PV, Parvovirus. The data underlying this figure can be found at the following DOI: https://zenodo.org/record/6968218#.Yu115vHMIUY.