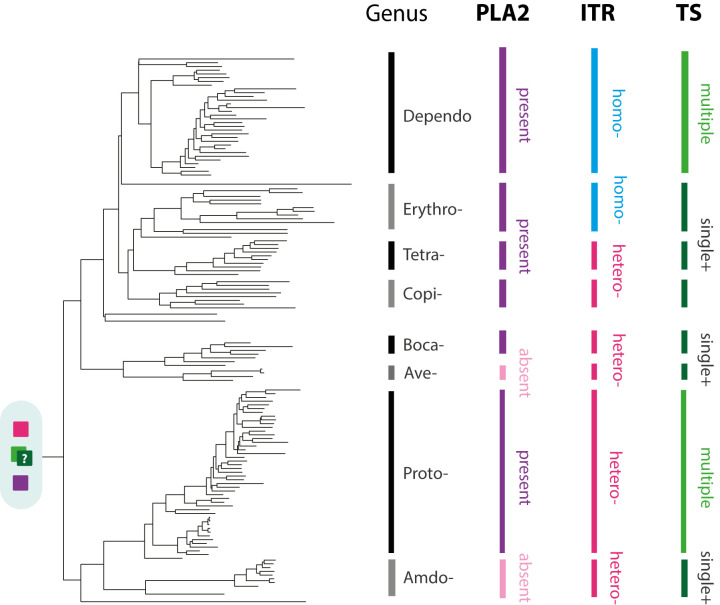
Fig 5. Conservation of genome features during Parvovirinae evolution.
A midpoint rooted, ML phylogeny showing the reconstructed evolutionary relationships between contemporary parvoviruses of subfamily Parvovirinae and the ancient parvovirus species represented by EPVs. The phylogeny shown here is shown in greater detail in Fig 4. The black and grey vertical bars to the right of the phylogeny indicate parvovirus genera. Coloured bars indicate the distribution of virus traits across genera, following the key. The most likely ancestral state is indicated at the root of the tree, based on the parsimonious assumption that independent losses of genome features are more likely than independent gains. The ancestral TS remains unclear. Abbreviations: EPV, endogenous parvoviral element; Hetero, heterotelomeric; Homo, homotelomeric; ML, maximum likelihood; MTSP, multiple transcriptional start positions; STSP+, single transcription start position, plus additional strategies; TS, transcription strategy. The data underlying this figure can be found at the following DOI: https://zenodo.org/record/6968218#.Yu115vHMIUY.