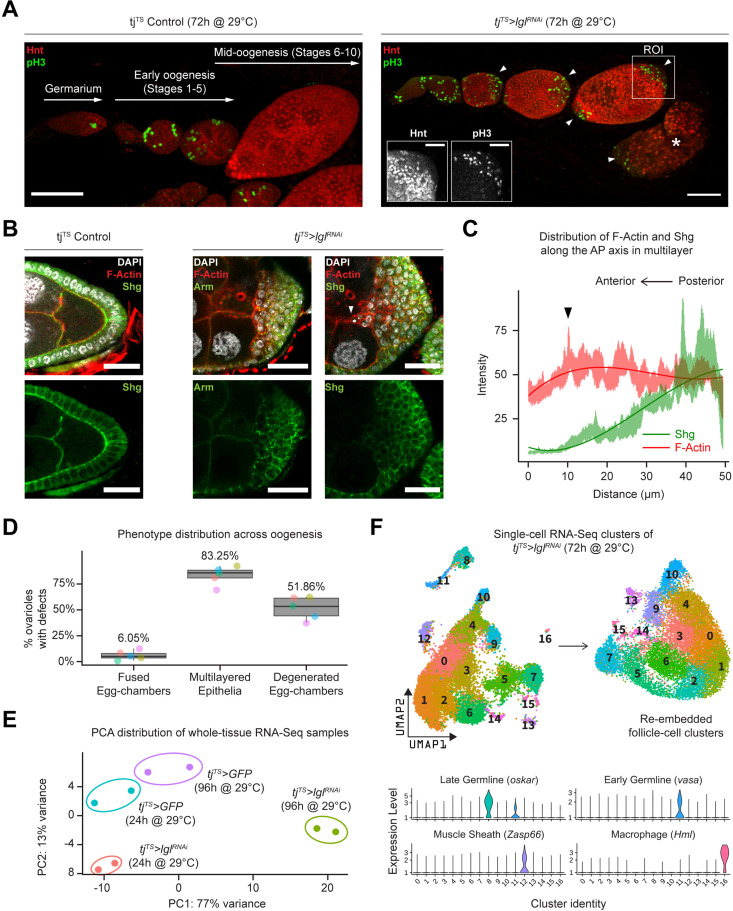
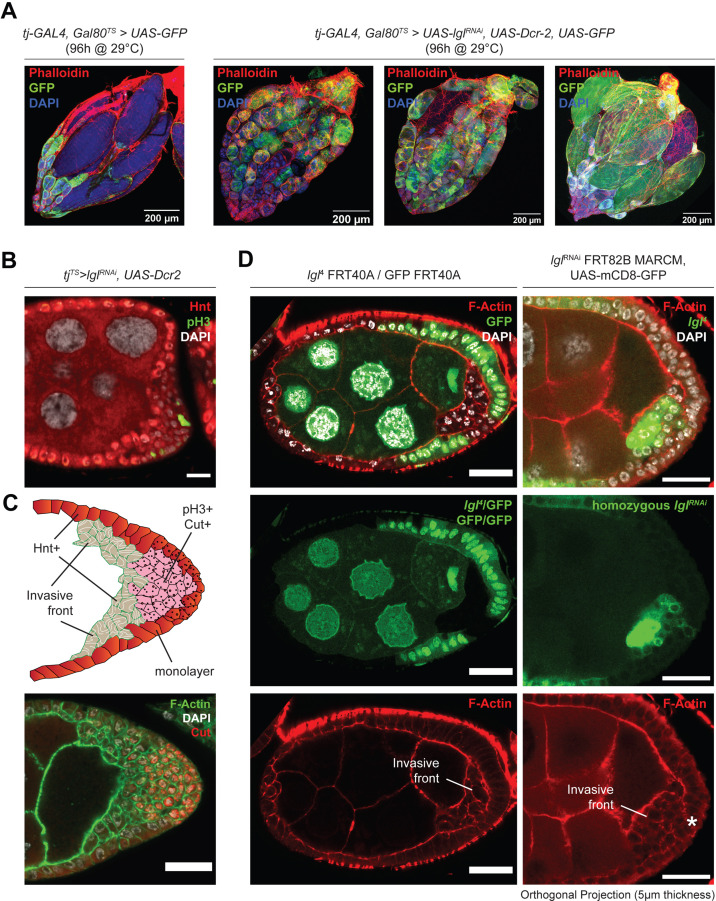
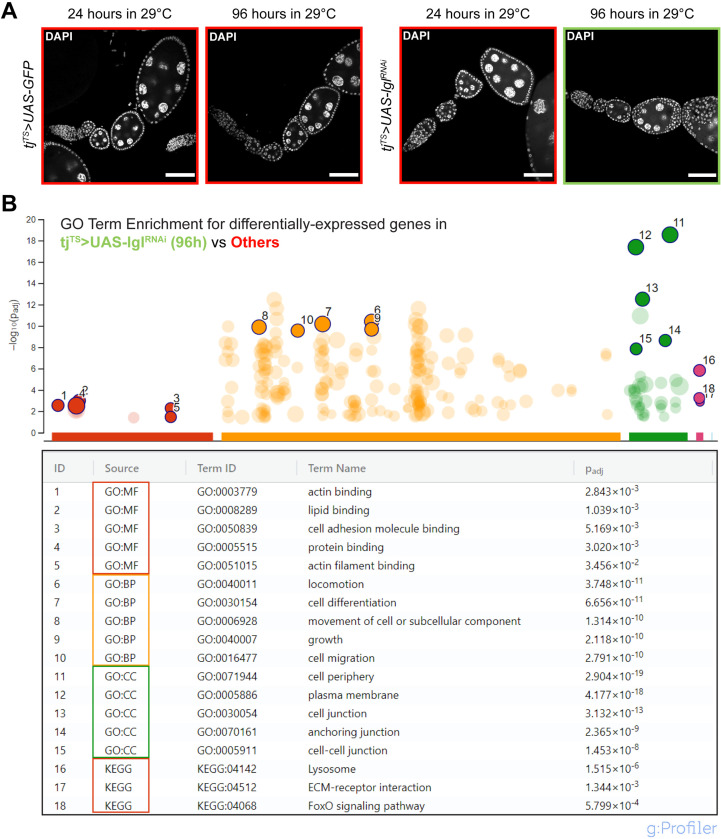
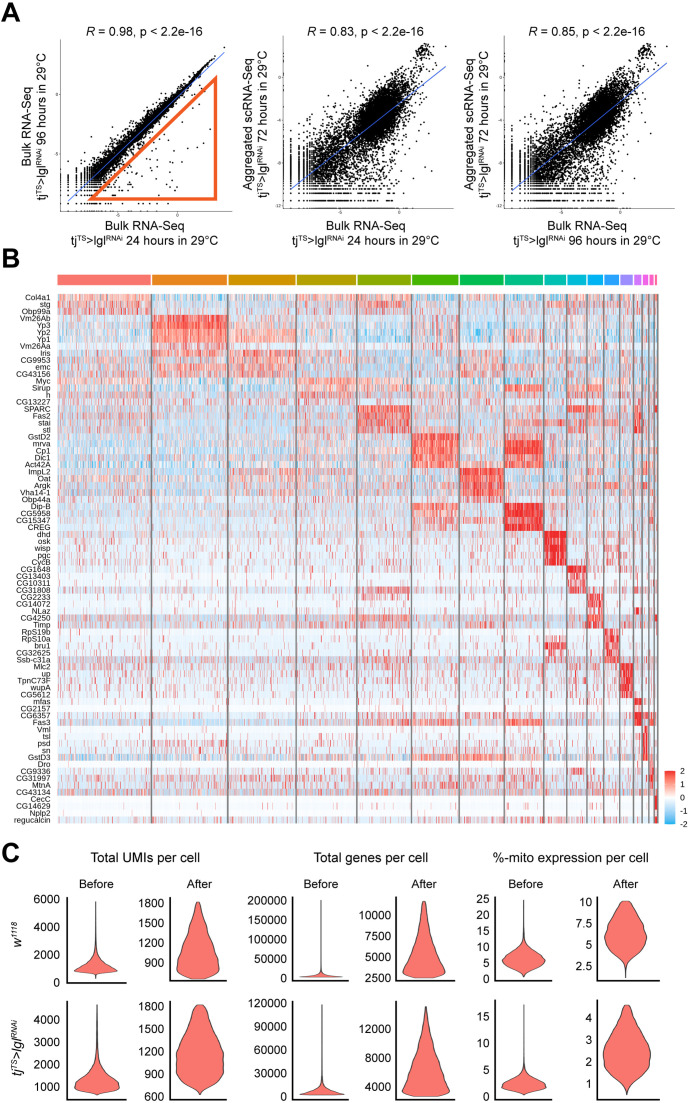
Figure 1. Inducing Lgl knockdown in follicle cells causes distinct phenotypic and transcriptomic changes.
(A) Left: orthogonal projection of a single ovariole displaying individual egg chambers containing experimental control follicle cells at early and midoogenesis. Follicle cells at mitotic stages are infrequently detected by pH3 staining (green), while endocycling follicle cells at midoogenesis are labeled by Hnt staining (red). Right: ovariole containing egg chambers with Lgl-KD in follicle cells exhibit continued cell division (marked by pH3 staining in green) in cells that accumulate at egg chamber termini (arrowheads) at early-to-midoogenesis developmental transition and midoogenesis. Degenerated egg chambers containing dying germline cells are marked by asterisks (*). Scale bars: 50 µm. Distinct Hnt and pH3 staining within the Lgl-KD multilayers are highlighted for the region of interest (ROI) within the image. ROI scale bar: 20 µm. (B) Left: cross-section of the posterior egg chamber epithelia containing experimental control follicle cells that exhibit intact Shg (DE-Cad) staining at cell junctions. Middle and right: posterior multilayers of egg chambers containing Lgl-KD in follicle cells show declining enrichment (green) of Armadillo (Arm; middle panel) and Shg (DE-Cad; right panel) along the anterior–posterior (AP) axis (right to left). F-actin (red) is found enriched in cells at the apical-most layers; leading edge of the invasive front is indicated by an arrowhead. Nucleus is marked by DAPI (white). Scale bars: 20 µm. (C) Relative enrichment of F-actin (red) and Shg (DE-Cad; green) along the AP axis across the biggest distance between the apical-most and basal-most cells in the multilayer shown in (B) (right panel). Intensities are measured across a 10 µm thickness (Z-axis) and a trendline (Gaussian fit) is shown. The black arrowhead marks the leading edge corresponding to that shown in (B). (D) Box-and-whisker plot showing quantification of the different tjTS>lglRNAi (72 hr in permissive temperature) phenotypes. Data was collected from five replicate trials (color-coded individually), consisting of 1250 intact ovarioles from a total of 165 flies. (E) Principal component analysis (PCA) plot showing the distribution of whole-tissue RNA-seq samples for tjTS experimental controls kept for 24 hr and 96 hr in permissive temperature and their experimental counterparts containing tjTS>lglRNAi. Each uniquely colored sample has two replicates that are grouped. (F) Overview of the single-cell (sc) RNA-seq workflow to isolate follicle cell-specific clusters from 14,537 tjTS>lglRNAi ovarian cells, embedded on lower UMAP dimensions (top). Clusters containing nonepithelial cell types are identified by the enrichment of specific markers (bottom).