FIGURE 3.
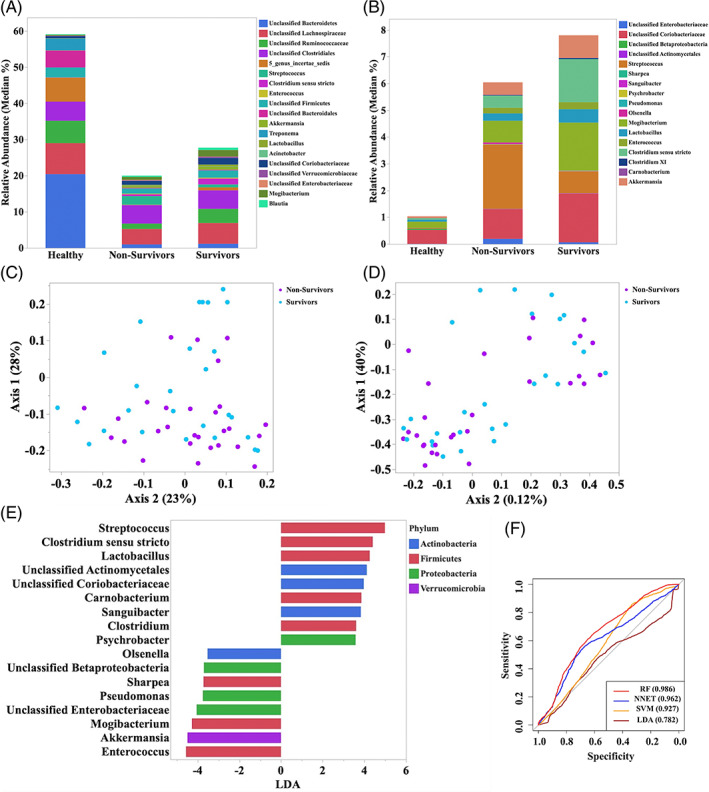
(A) Relative abundance (median) of the more abundant genera identified in surviving (n = 27) and nonsurviving (n = 28) horses with colitis. (B) Principal coordinates analysis (PCoA) based on Jaccard index analysis of the bacterial 16S rRNA gene sequence data for fecal samples collected from surviving (n = 27) and nonsurviving (n = 28) horses with colitis. (C) Principal coordinates analysis (PCoA) based on Yue and Clayton index analysis of the bacterial 16S rRNA gene sequence data for fecal samples collected from surviving (n = 27) and nonsurviving (n = 28) horses with colitis. (D) Relative abundance analysis of the genera identified in the LEfSe analysis to be enriched in surviving (n = 27) and nonsurviving (n = 28) horses with colitis. (E) Plot from LEfSe analysis indicating enriched taxa in fecal samples from surviving (n = 27) and nonsurviving (n = 28) horses with colitis. Linear discriminant analysis (LDA) cut off >3.5 and P < .05. (F) Area under the receiving operator curves of the random forest (RF), neural network (NNET), support vector machine (SVM), and linear discriminate analysis (LDA) for prediction of outcome (surviving versus nonsurviving) based on the fecal microbiota profile
