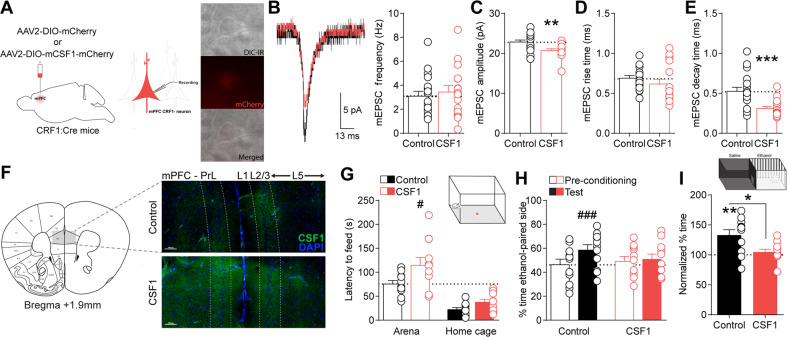
Fig. 5. CSF1 overexpression in mPFCCRF1+ neurons decreases post-synaptic glutamate transmission and is sufficient to increase anxiety-like behavior and abolish ethanol place preference.
A Schematic of viral strategy to selectively overexpress CSF1 in a Cre-dependent manner in mPFCCRF1+ neurons using ethanol-naïve CRF1:Cre mice. 40X magnification for brightfield, mCherry fluorescence, and merged images of a CSF1-mCherry expressing mPFCCRF1+ neuron. B Representative current trace of average mEPSC in control mCherry (black) and CSF1-mCherry (red) expressing mPFCCRF1+ neuron. Average mEPSC frequency in control mCherry and CSF1-mCherry expressing mPFCCRF1+ pyramidal neurons. C–E Average mEPSC amplitude, rise time, and decay time in control mCherry and CSF1-mCherry expressing mPFCCRF1+ neurons. n = 16–18 cells from N = 4 female mice; **p < 0.01, ***p < 0.001 by t-test. F Coronal mouse brain atlas image of mPFC and corresponding representative 10X magnification fluorescence image of boxed region depicting CSF1 expression in CSF1-overexpressing compared to control mice. Scale bar = 100 µM. G Latency to feed in an open arena (open bars) and home cage (filled bars) in the novelty-suppressed feeding test in control (black) and mPFCCRF1+ CSF1 overexpressing (red) mice with significant main effects of condition F(1,19) = 58.59, p < 0.001 and group F(1,19) = 5.67, p = 0.02 by two-way ANOVA, and post hoc significance #p < 0.05 is represented in panel. H Percent time spent in the ethanol paired context during the pre-conditioning phase (open bars) and following ethanol conditioning (filled bars) in the ethanol place conditioning test for control (black) and mPFCCRF1+ CSF1 overexpressing (red) mice with a significant main effect of condition F(1,20) = 11.43, p = 0.003 and interaction effect F(1,20) = 6.41, p = 0.01 by two-way ANOVA, and post hoc significance ###p < 0.001 is represented in panel. I Pre-conditioning normalized percent time spent in the ethanol-paired side with significant **p < 0.01 effects by one-sample t-test and *p < 0.05 unpaired t-test. N = 11 female mice/group.