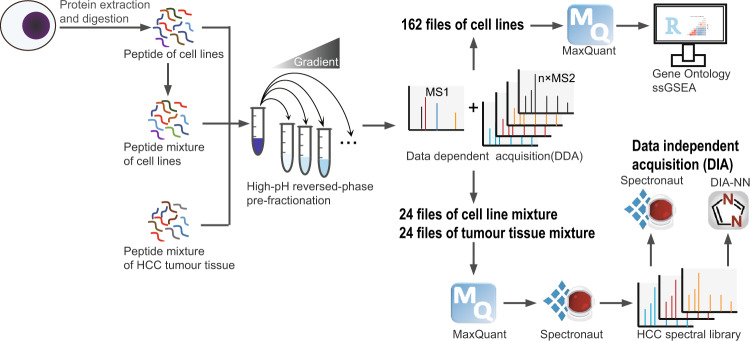
Fig. 1.
Workflow for the proteomic analysis of nine HCC cell lines and generation of the HCC spectral library. Nine HCC cell lines were protein extracted and trypsin digested. Peptides were pre-fractionated by Hp-RP and analysed by LC-MS/MS using DDA mode. The gained 162 raw files were searched again protein database by MaxQuant version 2.0.3.0. Gene Ontology (GO) and ssGSEA analysis were then implemented. For generation of the HCC spectral library, peptides of HCC cell line mixture or tumour tissue mixture were pre-fractionated, then analysed by LC-MS/MS using DDA mode. The gained 48 raw files were searched again protein database by MaxQuant version 2.0.3.0, and the search results were imported into SpectronautTM version 15.2 to generate the HCC spectral library. The HCC spectral library could be used in SpectronautTM version 15.2 and DIA-NN version 1.8 for DIA quantification.