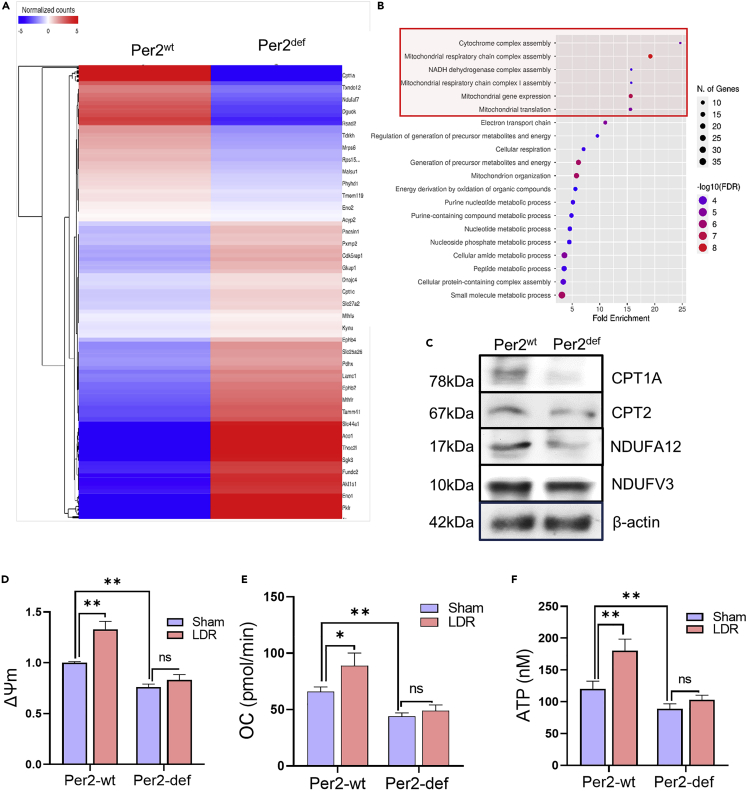
Figure 3.
Per2 is required for LDR-induced mitochondrial activation (see also Figure S4)
(A) A cluster of genes related to mitochondrial metabolic functions by RNAseq analysis of Per2wt versus Per2def BM-HSCs with 1.2-fold cutoff.
(B) Gene ontology biological process enrichment analysis of up-regulated genes related to mitochondrial metabolic functions with 1.2-fold cutoff in Per2wt versus Per2def BM-pHSCs.
(C–F) Western blot of a cluster of mitochondrial metabolic factors CPT1A, CPT2, NDUFA12, and NDUFV3 in Per2wt and Per2def BMMNCs. Mitochondrial membrane potential (D), oxygen consumption (E), and ATP generation (F) were measured in Per2wt and Per2def BMMNCs 24 h after LDR.
Data are represented as mean ± SEM, n = 3, ∗p < 0.05; ∗∗p < 0.01, ns p > 0.05, Student’s t test.