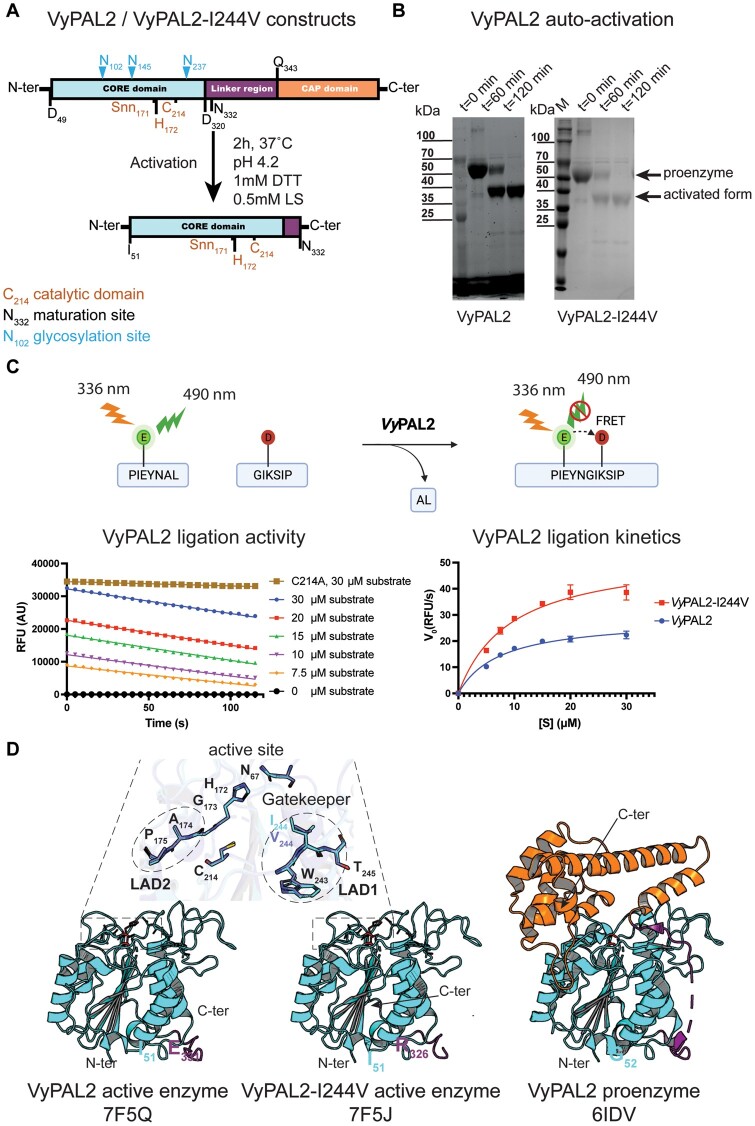
Figure 2.
Expression, activation, ligase activity, and crystal structure of VyPAL2-I244V. A, Schematic sequence representation of VyPAL2-I244V. The respective VyPAL2 genes encoding complete amino-acid sequences were cloned into the expression vector, with the signal peptide substituted by a hexa-His-tag for affinity purification (see “Materials and methods”). Catalytic residues and the aspartimide moiety (Snn171) next to the catalytic His172 are indicated in brown, N-linked glycosylation sites are indicated in cyan, and domain boundaries (as deduced from a previous LC–MS/MS analysis) are indicated in black (please also see SI of Hemu et al. (2019)). Proteolytic cleavage sites are indicated with arrows above the N- and C-terminal sequences. The products from low pH activation of the proenzyme were analyzed using (B) SDS–PAGE with Coomassie blue staining. Incubation of VyPAL2 for 2 h led to the production of a fully mature enzyme. C, Analysis of ligase activity using a Förster Resonance Energy Transfer assay. Red squares (VyPAL2-I244V) and blue points (VyPAL2) represent mean values and error bars denote standard deviations. D, Comparison between the crystal structures of the VyPAL2 pro-enzyme monomer (right, PDB access code: 6IDV (Hemu et al., 2019)) and the activated VyPAL2 protein (left, this work). The proteins are displayed with α-helices as ribbons and β-strands as arrows. The color-code used for each of the three domains of the proenzymes is used throughout the manuscript (core domain: green, cap domain: wheat (light yellow), linker region connecting the catalytic core and cap domain: orange).