Figure 6.
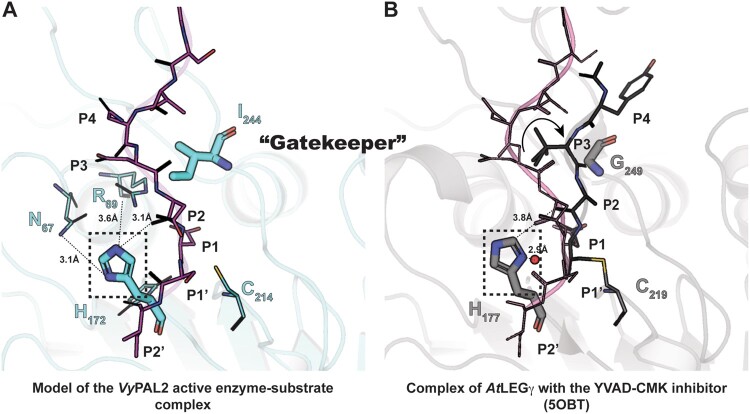
Active site residues and the role of the Gatekeeper position in substrate positioning. A, Structure of the enzyme–substrate complex simulated based on the experimental VyPAL2-C214A structure (Supplemental Table S2, form I). Ala214 was replaced with a cysteine. The enzyme core is shown as ribbons and colored in slate. The catalytic Cys214 and His172 as well as the Ile244 Gatekeeper residue are shown as sticks. Residues Asn67 and Arg69 putatively involved in the auto-activation mechanism are shown as sticks with distances from the His172 imidazole ring indicated (see text). The hydrogens of His172 are also shown. The P4–P2′ (NDDDSI) substrate is shown as sticks, colored in magenta, and the 2Fo–Fc electronic density map contoured at 1σ is colored in light blue. B, Structure of the enzyme–inhibitor complex of AtLEGg (PDB code: 5OBT). The enzyme core is shown as ribbons and colored in light gray. The catalytic residues Cys219 and His172 as well as the Gly249 Gatekeeper residue are shown as sticks. The YVAD-CMK covalent inhibitor is shown as sticks, with residues labeled P4–P1. The proposed nucleophilic water molecule (shown as a red sphere) is located at a distance of 2.9 Å from His172 in the axis of the thioester bond between the Cys219 and P1 Asp–CMK residues. For reference, the position of the VyPAL2-C214A substrate superimposed onto the AtLEGγ bound structure is represented as lines and colored in magenta.