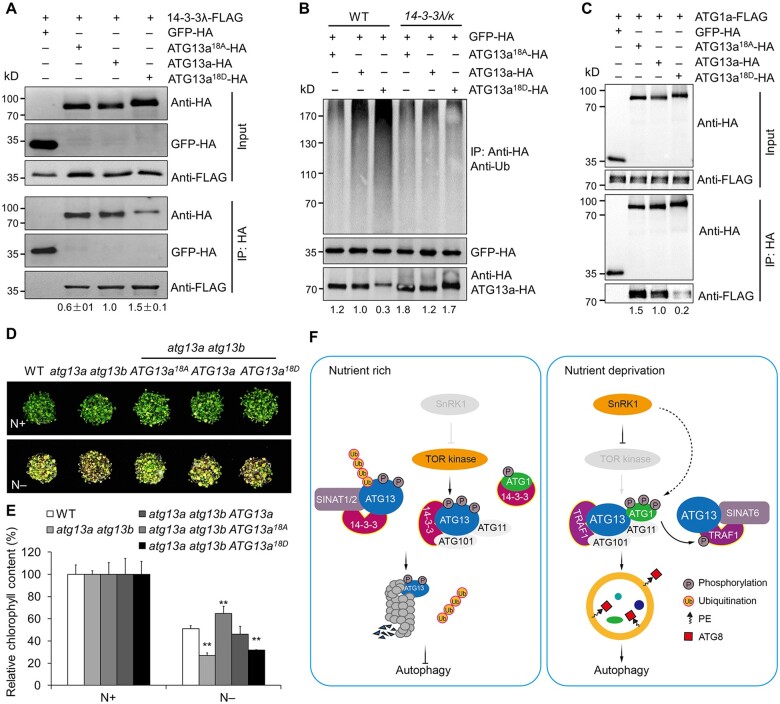
Figure 7.
14-3-3s interact with phosphorylated ATG13a to modulate ATG13a degradation and ATG1a–ATG13a complex formation. A, In vivo Co-IP assay of the interaction between 14-3-3λ and ATG13a with different phosphorylation states. Constructs encoding 14-3-3λ-FLAG and HA-tagged ATG13a in different phosphorylation states (ATG13a18A-HA, ATG13a-HA, or ATG13a18D-HA) were transiently co-transfected into protoplasts from WT plants and incubated under light conditions for 16 h, before immunoprecipitation with HA affinity agarose beads. The blots were probed with anti-HA and anti-FLAG antibodies. Relative intensity of the immunoprecipitated 14-3-3λ-FLAG normalized to the different phosphorylation states of ATG13a is shown below. The relative intensity of each protein is the mean ± sd (n = 3) calculated from three independent experiments. B, Ubiquitination of ATG13a in different phosphorylation states in the 14-3-3λ 14-3-3k double mutant. Constructs encoding HA-tagged ATG13a in different phosphorylation states (ATG13a18A-HA, ATG13a-HA, or ATG13a18D-HA) were transiently transfected in Arabidopsis protoplasts isolated from 4-week-old WT and 14-3-3λ 14-3-3k mutant plants and incubated for 16 h under continuous light conditions before the ubiquitination and stability of ATG13a was determined by immunoblot analysis using anti-Ub and anti-HA antibodies. The relative intensity of each protein normalized to GFP-HA is shown below. C, Interaction between ATG13a in different phosphorylation states and ATG1a by Co-IP assay. Constructs encoding FLAG-tagged ATG1a (ATG1a-FLAG) and HA-tagged ATG13a in different phosphorylation states (ATG13a18A-HA, ATG13a-HA, or ATG13a18D-HA) were transiently co-transfected into protoplasts from WT plants and incubated under light conditions for 16 h. Total proteins were extracted and immunoprecipitated with HA affinity agarose beads; the blots were probed with anti-HA and anti-FLAG antibodies. Numbers on the left indicate the molecular weight (kDa) of each band. The relative intensity of the immunoprecipitated ATG1a-FLAG normalized to the different phosphorylation states of ATG13a is shown below. D, Sensitivity of the WT, atg13a atg13b double mutant, and atg13a atg13b ATG13a, atg13a atg13b ATG13a18A, atg13a atg13b ATG13a18D transgenic lines to N starvation. Seedlings were grown for 1 week on MS medium, followed by transfer to N-rich (N+) or N deficient (N–) medium for an additional 4 days. E, Relative chlorophyll contents of seedlings with or without N starvation shown in (D). The relative chlorophyll contents were calculated by comparing the values of N–treated versus N+-treated seedlings. Data are means ± sd (n = 3) calculated from three independent experiments. For each experiment, three technical replicates from pools of 15 seedlings each were used per genotype. Asterisks indicate significant differences from the WT (**P < 0.01 by Student’s t test). F, Working model for 14-3-3 family proteins in regulating autophagy in Arabidopsis. Under nutrient-rich conditions, the TOR kinase complex is active and phosphorylates ATG13a to maintain it in a hyperphosphorylated state. The 14-3-3 proteins associate with the phosphorylated form of ATG13a and act as adaptors to recruit the E3 Ub ligases SINAT1 and SINAT2 to form a protein complex that promotes the ubiquitination and degradation of ATG13 via the 26S proteasome pathway. Phosphorylated ATG13 also disassociates from ATG1, suppressing autophagy. Under nutrient starvation, however, SnRK1 inhibits TOR activity, resulting in ATG13 dephosphorylation, which impairs interaction with 14-3-3 proteins and forms a complex with hyperphosphorylated ATG1a mediated by the SnRK1. The scaffold proteins TRAF1s interact with ATG13a together with SINAT6, forming a protein complex that decreases SINAT1- and SINAT2-mediated ubiquitination and degradation of ATG13 to induce autophagy. Furthermore, the ATG1-mediated phosphorylation of TRAF1s in vivo may represent a positive feedback loop that promotes the stability and function of TRAF1 proteins in autophagy induction.