Extended Data Fig. 6|. Integrated ChIP-seq and RNA-seq analysis showing the EZH2:PRC2 on-target effect of MS177.
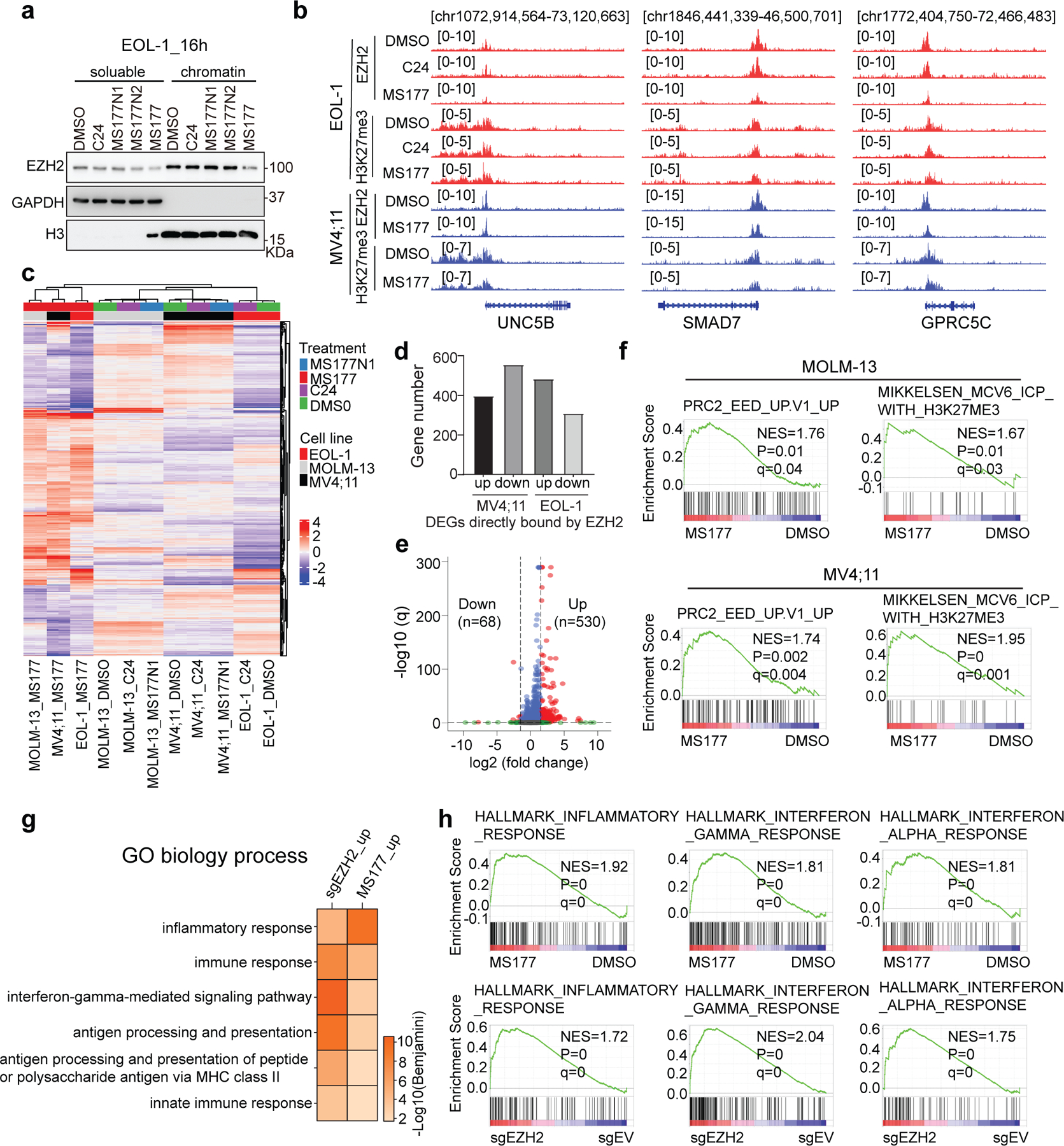
(a) Immunoblotting of EZH2, either nucleoplasmic (left) or chromatin-bound (right), in EOL-1 cells after a 16-hour treatment with DMSO or 2.5 µM of C24, MS177N1, MS177N2 or MS177. GAPDH and histone H3 serve as the cell fractionation controls.
(b) IGV views of EZH2 and H3K27me3 ChIP-seq peaks at the indicated EZH2:PRC2 target gene post-treatment of EOL-1 (upper; for 16 hours) and MV4;11 cells (bottom; for 24 hours) with either DMSO or 0.5 μM of C24 or MS177.
(c) Unsupervised clustering analysis using the RNA-seq-based transcriptome profiles of the three independent MLL-r acute leukemia cell lines after the treatment with DMSO or 0.5 μM of MS177, C24 or MS177N1. MV4;11 and MOLM13 cells were treated for 24 hours, and EOL-1 cells for 16 hours (n=2 replicated samples).
(d) Bar plot showing the number of DEGs associated with the direct EZH2 binding in MV4;11 (left) or EOL-1 (right) cells. Up and down refer to those up- and down-regulated DEGs, respectively, based on RNA-seq analysis.
(e) Volcano plot showing differential expression analysis of genes based on RNA-seq profiles of EOL-1 cells with EZH2 KO versus mock control (n=2 replicated samples). The x-axis shows the log2 value of fold-change in gene expression (in KO versus vector-treated cells) and the y-axis shows the -log10 value of adjusted P (q) value, with the dashed lines indicating the cut-off of significance.
(f) GSEA revealing that, relative to DMSO, MS177 treatment is positively correlated with upregulation of the indicated genes repressed by PRC2:EED (left) or bound by H3K27me3 (right) in MOLM-13 (top) or MV4;11 (bottom) cells.
(g) DAVID functional annotation reveals that the DEGs up-regulated after EZH2 KO (sgEZH2_up) or MS177 treatment (MS177_up) in EOL-1 cells have similar enrichment for the immunity-related genes.
(h) GSEA shows that, relative to their respective controls, both MS177 treatment (top) and EZH2 KO (bottom) in EOL-1 cells are positively correlated with upregulation of the indicated immunity-related genesets.
