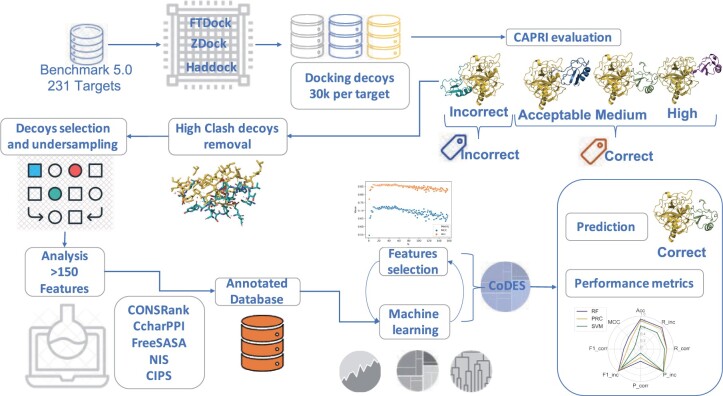
Fig. 1.
An overview of the classifier development. docking decoys were generated for the BM5 targets with FTDock, ZDOCK and HADDOCK. Based on the CAPRI quality criteria, they were labeled as correct or incorrect; where the ‘correct’ class includes the acceptable, medium- and high-quality ones. Decoys with a high number of clashes were removed. By undersampling, balanced and unbalanced (in terms of included correct and incorrect decoys) datasets were derived for training and validation. For each decoy, 158 features (scoring functions and complex descriptors) were calculated. Three different ML algorithms were applied, from which, upon validation, the RF one was selected. The optimized RF classifier, CoDES, was then independently tested