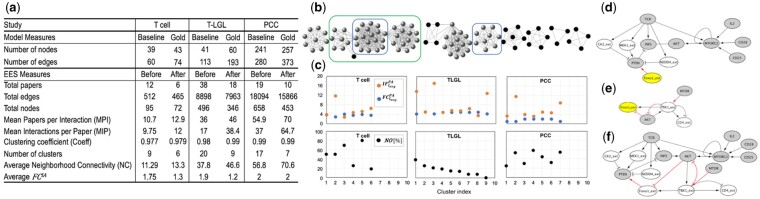
Fig. 3.
(a) Description of each use case in terms of the size of both baseline and gold standard models, followed by the values of several graph metrics for the ECLG before and after the removal of less frequent events, for T-cell, T-LGL and PCC case studies. (b) Candidate ECLG for the T-cell case study. (c) (Top) Average literature support metrics and for generated clusters. (Bottom) Node overlap (NO) between the clusters and the baseline model for the three case studies. (d–f) Cluster interpretation and preparation for extension: (d), (e) Interpreted clusters’ influence graphs, C3, C2, respectively, for T-cell case study, nodes are biological entities, pointed arrows represent activation, blunt arrows represent inhibition. Baseline model nodes are in gray and the new nodes with suffix ‘_ext’ are in white. (d) Cluster C3 with upstream element Foxo1_ext highlighted in yellow, (e) cluster C2 with downstream element Foxo1_ext. The return path from C2 to C3 is (‘MTOR—TBK1_ext—AKT– Foxo1_ext—PTEN’), highlighted in red, the first node and last node of the path are MTOR and PTEN, respectively, (f) the result of merging C3 and C2