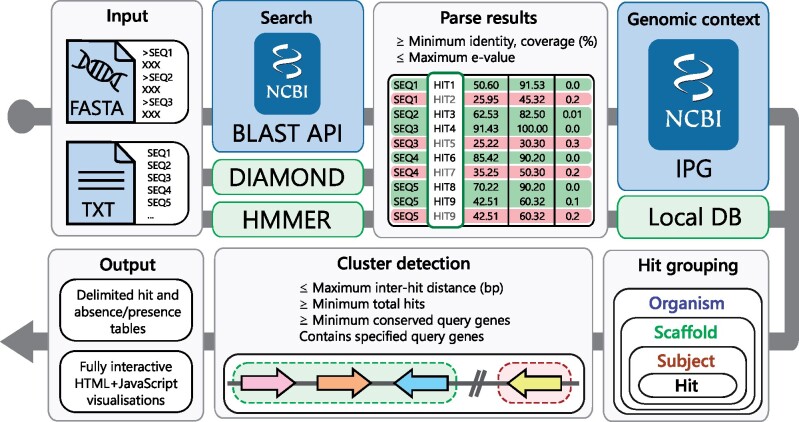
Fig. 1.
The cblaster search workflow. Input sequences are given either as a FASTA file or as a text file containing NCBI sequence accessions. They are then searched against the NCBI’s BLAST API or a local DIAMOND database, in remote (blue background) and local (green background) modes, respectively. BLAST hits are filtered according to user-defined quality thresholds. Genomic coordinates for each hit are retrieved from the IPG resource. Hits are grouped by their corresponding organism, scaffold and subjects. Finally, hit clusters are detected in each scaffold and results are summarized in output tables and visualizations