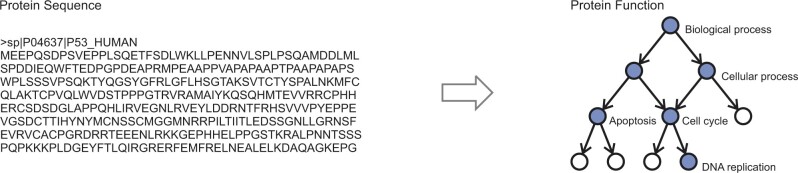
Fig. 1.
Stylized representation of the protein function prediction problem. (Left) Protein sequence of the human cellular tumor suppressor protein p53 consisting of 393 amino acids. The protein is shown in a FASTA format. (Right) Protein function defined as a set of blue nodes in the ontology graph consisting of 10 nodes. Each node is associated with a textual descriptor (term) from a controlled vocabulary; e.g. ‘biological process’ as the root, ‘apoptosis’ as an internal node, and ‘DNA replication’ as a leaf node. The terms in the ontology are connected by relational ties such as is-a, which are read in the direction opposite the arrow. The task for a function prediction algorithm is to take an input sequence and output the nodes (terms) colored in blue. Note that the prediction of ontological terms is a somewhat simplified view of function prediction as it, for the most part, ignores the contextual, spatio-temporal and causal relations