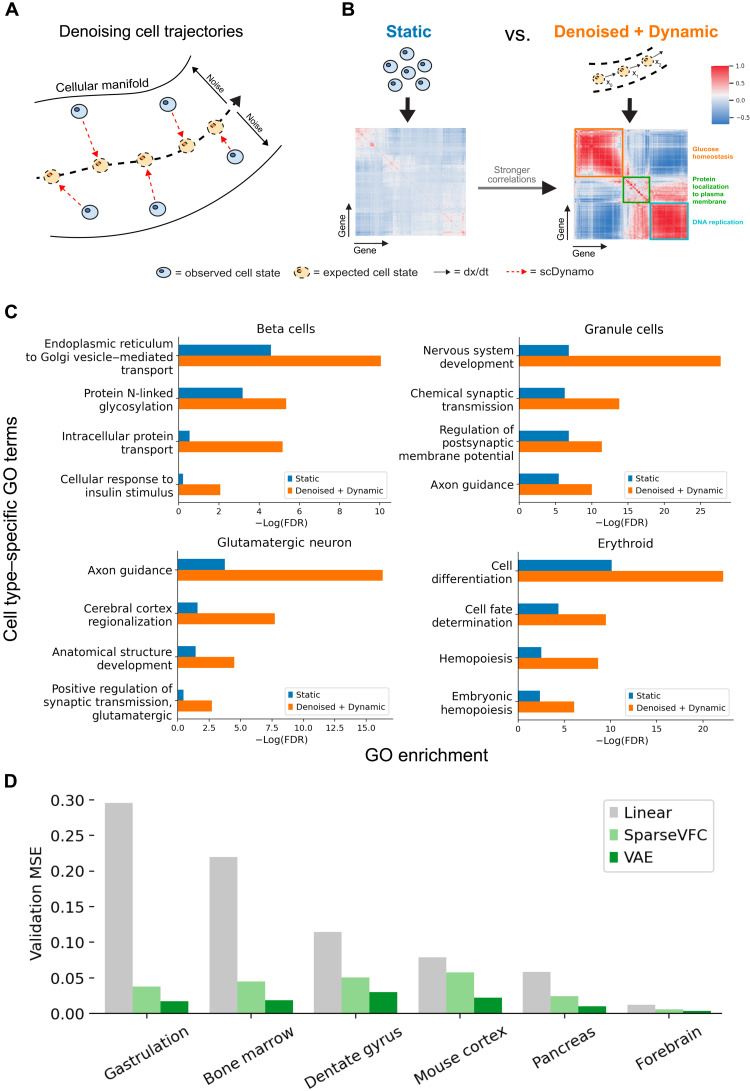
Fig. 5. Disentangling trajectory-specific gene coexpression networks.
(A) Schematic for reducing variability along a developmental trajectory due to sparsity and noise in scRNA-seq experiments with denoising VAEs in DeepVelo. (B) We used a representative initial condition (e.g., the median expression profile for a cluster of cells) to simulate denoised cell trajectories according to the dynamics learned by DeepVelo. Compared with static cell clusters, the dynamic cells in denoised trajectories have stronger correlations between genes, which leads to better coherence between functional gene modules. Given the same set of genes, the two correlation matrices are each biclustered to identify functional gene coexpression modules. (C) GO enrichment of cell type–specific terms from the most significant gene cluster. Gene coexpression modules from dynamic cells have higher cell type–specific GO term enrichment than gene modules found from existing methods. (D) Benchmarking with the linear and state-of-the-art vector-field learning method SparseVFC shows that our VAE-based framework can improve out-of-sample velocity vector prediction accuracy by at least 50% across the six datasets, indicating that DeepVelo can learn a more accurate representation of the velocity field.