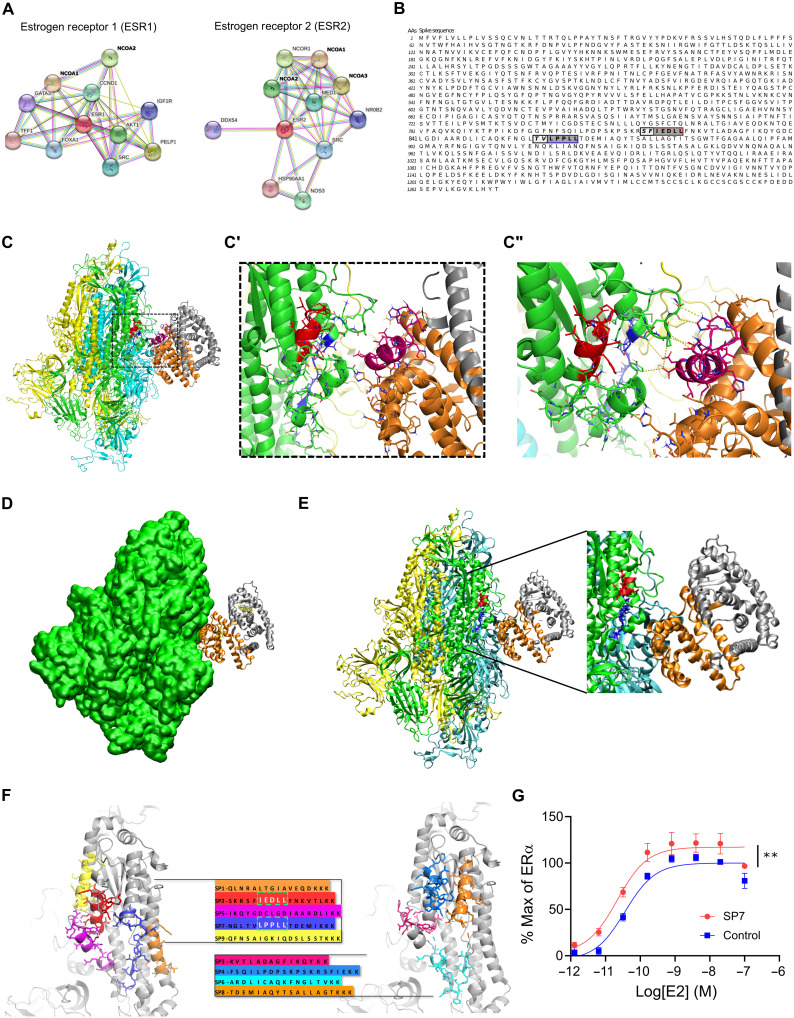
Fig. 2. S and ER interact at conserved LXD NRC motifs.
(A) ER interaction network showing known and predicted protein associations. (B) LXD-like patterns in the S sequence. The LXXLL motif and a homologous region are highlighted in blue and red boxes, respectively, with dark gray background. Positions (−1 and −2) are reported in italic and light gray background, respectively. AAs, amino acids. (C and C′) The LPPLL and IEDLL residues of the two motifs are shown in the 3D x-ray S structure [Protein Data Bank (PDB) ID 6VYB] with blue and red colors, respectively. The ER dimer is in orange and gray, while the helix-12 is reported in magenta. (C″) The image shows favorable interactions between ER and the S’s regions containing the LXXLL motifs. The interacting residues and the predicted interactions are reported in stick and yellow dots, respectively. (D) S-ER motif–oriented docking. The best 3D docking hypothesis is shown. The ER dimer is in orange and gray, and S is green. (E) Alignment between the best-pose and the 3OLL model tied with NCOA1. The region occupied by S’s α helix interacts in the area where the NCOA fragment was crystallized. (F) S protein peptides and their location with respect to the S 3D structure. (G) The SP7 peptide containing the LPPLL motif significantly increased ERα activation [F(1,48) = 30.38, **P < 0.01, two-way analysis of variance (ANOVA); peptide treatment main effect]. Data are shown as means ± SEM.