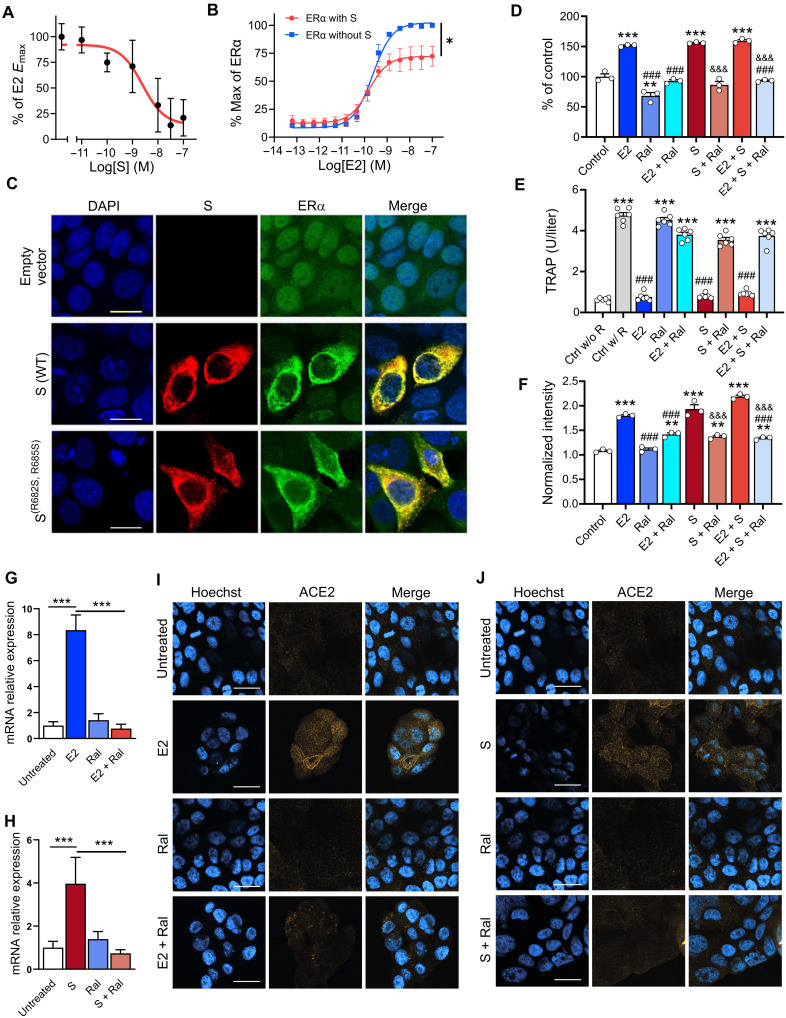
Fig. 3. S modulates ER-dependent biological functions.
S inhibits (A) E2-induced ERα DNA binding in MCF-7 nuclear extracts and (B) transcriptional activation in an ERα reporter cell line [F(1,28) = 21.73, *P = 0.01, two-way ANOVA; S treatment × E2 concentration interaction effect]. (C) Immunofluorescent staining of S and endogenous ERα in MCF-7 cells transfected with empty vector, WT, or the furin cleavage site mutant S(R682S, R685S). Scale bars, 16 mm. (D) S increases MCF-7 cell proliferation in an ER-dependent manner (**P < 0.01 and ***P < 0.001 versus control; ###P < 0.001 versus E2; &&&P < 0.001 versus S; one-way ANOVA with Tukey post hoc test). (E) S decreases osteoclast differentiation in an ER-dependent manner (***P < 0.001 versus control without RANKL; ###P < 0.001 versus control with RANKL; one-way ANOVA with Tukey post hoc test). (F) S and E2 increase ACE2 protein levels in MCF-7 cells in an ER-dependent manner (***P < 0.001 versus control; ###P < 0.001 versus E2; &&&P < 0.001 versus S; one-way ANOVA with Tukey post hoc test). (G and H) E2 and S increase ACE2 mRNA (***P < 0.001, one-way ANOVA with Tukey post hoc test) and (I and J) protein in the Calu-3 lung cell line in an ER-dependent manner. Scale bars, 30 mm. All data are shown as means ± SEM.