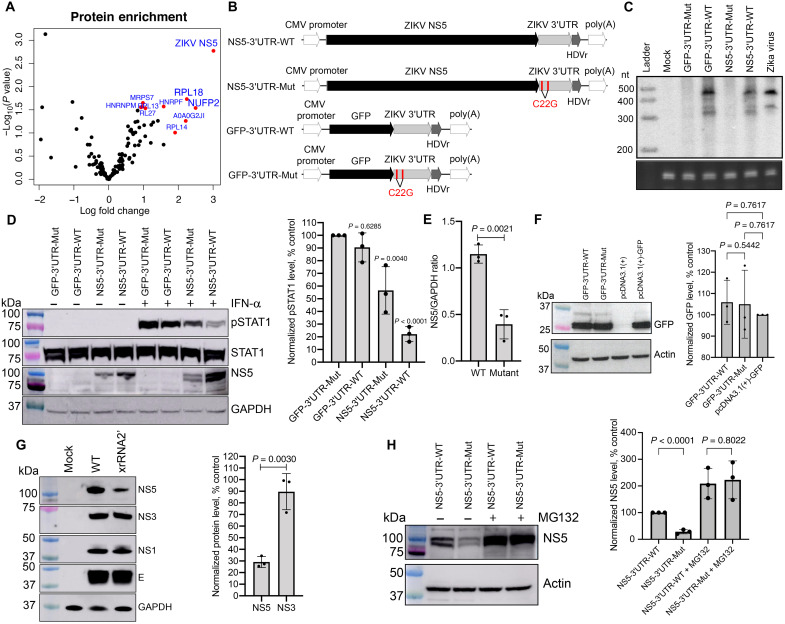
Fig. 8. ZIKV sfRNA inhibits STAT1 phosphorylation by stabilizing NS5 via direct binding.
(A) The sfRNAZIKV-binding proteins identified by RNA affinity pulldown. Top 10 most significant sfRNA-interacting proteins are highlighted. The values are the means from three independent experiments. (B) Schematics of the reporter constructs for the production of ZIKV sfRNA and NS5 alone and in combination. The mutations leading to sfRNA deficiency are shown in red. CMV, cytomegalovirus; poly(A), polyadenylate. (C) Production of sfRNA in HEK293T cells transfected with the constructs shown in (B). Bottom: 5.8S rRNA. (D) Detection of pSTAT1, total STAT1, NS5, and GAPDH (loading control) in HEK293T cells transfected with the plasmids shown in (B) and treated with IFN at 48 hours posttransfection (hpt). (E) Quantification of NS5 levels in (D) by densitometry. (F) Western blot for GFP expression in HEK293T cells transfected with the plasmids expressing GFP with and without sfRNAZIKV. (G) Detection of structural (E-protein) and nonstructural (NS5, NS3, and NS1) proteins in Vero cells infected with WT and xrRNA2′ ZIKV. (H) Effect of the proteasome inhibitor MG132 on the accumulation of ZIKV NS5 in HEK293T cells 24 hours after transfection with indicated constructs. Right panels in (D) and (F) to (H) shows image densitometry for the respective blots based on three independent experiments. Values are normalized to GAPDH or actin and expressed as a percentage relative to control. The blots in (C), (D), and (F) to (H) are representative of three independent experiments. Error bars in (D) to (H) represent SD. Statistical analysis was performed by one-way ANOVA (E) or t test (E to H).