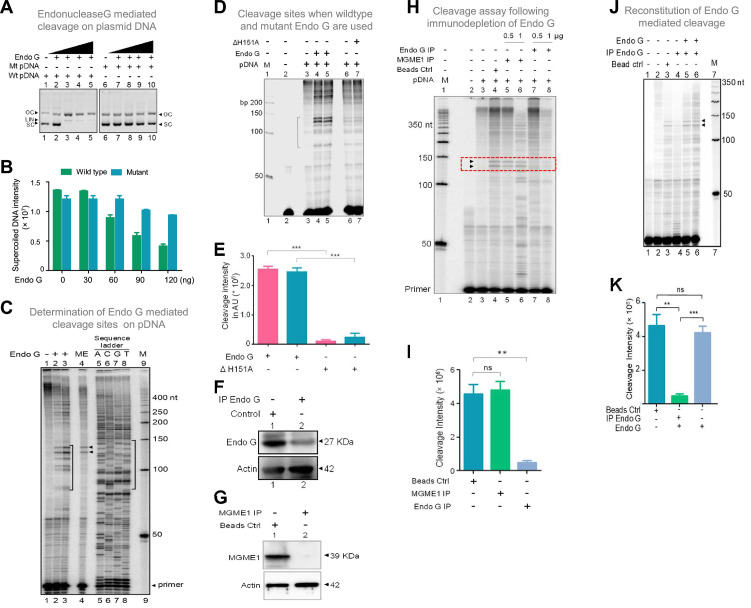
Figure 7. Studies to identify mitochondrial nuclease responsible for cleavage at G4 DNA formed at Region I of the mitochondrial genome.
(A) In vitro nicking assay using purified Endonuclease G on wild type and mutant plasmids containing mitochondrial Region I (pDI1 and pDI2). Both wild type and mutant plasmids were treated with increasing concentration of purified Endonuclease G and resolved on a 0.8% agarose gel. Lanes 1 and 6 served as the control without any protein in the reaction whereas lanes 2–5 and lanes 7–10 are with increasing concentration (30, 60, 90, 120 ng) of purified Endonuclease G. (B) Quantification showing the efficiency of Endonuclease-G-mediated cleavage when a plasmid containing mutant and wild type G4 motif derived from Region I was compared. (C) Cleavage assay was performed on a plasmid containing mitochondrial Region I, pDI1 following incubation with purified Endonuclease G. Primer extension assay was carried out using [γ-32P] radiolabeled VKK11 primer and resolved on 8% denaturing polyacrylamide gel. Lanes 2 and 3 represent 30 and 60 ng of Endonuclease G incubated samples. Lane 1 is without protein, lane 4 is with mitochondrial extract (ME). and lanes 5–8 are A, C, G, T represents the sequencing ladder. ‘M’ is 50 bp ladder. Marked regions represent the specific cleavage products. (D) Cleavage assay was performed on a plasmid containing mitochondrial Region I, pDI1 following incubation either with purified Endonuclease G or mutant Endonuclease G (ΔH151A). Primer extension assay was carried out using γ-32P radiolabeled VKK11 primer and resolved on 8% denaturing polyacrylamide gel. Lane 2 is primer alone, Lanes 3 and 6 are without protein, lanes 4 and 5 are with 30 and 60 ng of Endonuclease G, lanes 7 and 8 are with 30 and 60 ng of mutant Endonuclease G incubated samples. ‘M’ is 50 bp ladder. (E) Quantification showing the efficiency of wild type and mutant Endonuclease G (ΔH151A) mediated cleavage when a plasmid containing wild type G4 motif derived from Region I was compared. (F) Western blotting showing immunodepletion of Endonuclease G from rat testicular mitochondrial extracts. Protein A/G beads were incubated with anti-Endonuclease G and then with the extracts. Actin served as a loading control. (G) Immunodepletion of another endonuclease present in mitochondria, MGME1 from rat testicular mitochondrial extracts as described in panel F. (H) Endonuclease G or MGME1-depleted extract was incubated with pDI1 and used for the primer extension using VKK11 primer. Extract without the addition of antibody served as bead control (lane 4). Lane 3 is no protein control, lanes 5 and 6 corresponds to increasing concentrations of MGME1 depleted extracts, while in lanes 7 and 8, increasing concentrations of Endonuclease G depleted extracts were added. ‘M’ is 50 nt ladder. Cleavage positions are indicated by arrow and boxed (red). (I) Bar diagram depicting quantitation showing the impact of immunoprecipitation of Endonuclease G based on multiple experiments. (J) Reconstitution assay was performed by the addition of purified Endonuclease G following its immunodepletion. Lane 2 represents the primer alone; Lane 3 represents the beads control; lane 4 represents the Endonuclease G depleted extract while lanes 5 and 6 represent the addition of purified Endonuclease G to the depleted extracts (performed in duplicate reaction). (K) Bar diagram representing the cleavage intensity after reconstitution assay as shown in panel J. In panels, C, D, H and J, ‘M’ represents 50 nt ladder. In panels E, I and K, quantitation is based on three biological repeats and data is shown with error bar calculated as mean ± SEM (ns not significant, *p<0.05, **p<0.005, ***p<0.0001). Refer also Figure 7—figure supplement 1.