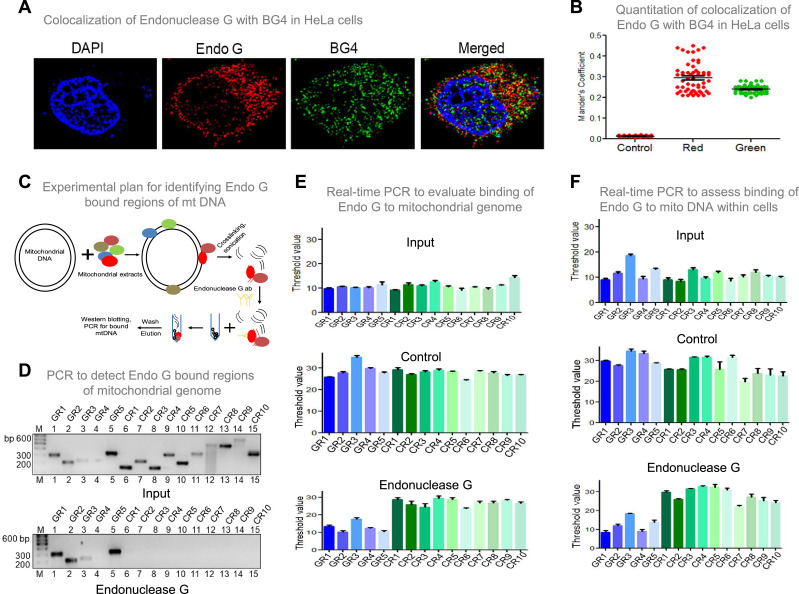
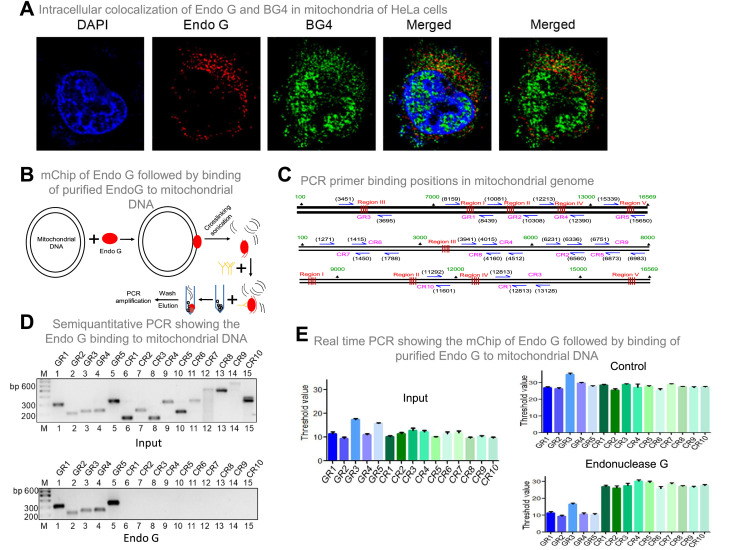
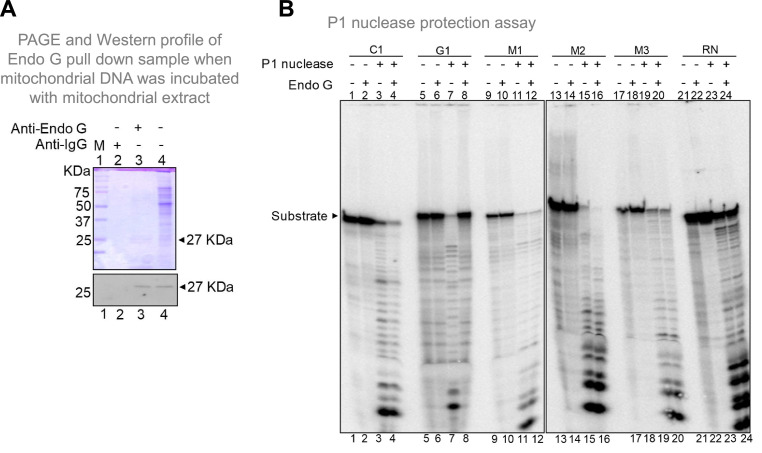
Figure 9. Investigation of binding efficacy of Endonuclease G to G4 DNA at Region I of mitochondrial genome.
(A) Representative image showing colocalization of Endonuclease G with BG4 in HeLa cells. Alexa Fluor 568 and Alexa Fluor 488 conjugated secondary antibodies were used for detection of Endonuclease G and BG4 proteins, respectively. DAPI was used as nuclear stain. (B) The quantitation showing colocalization of Endonuclease G and BG4. The colocalization was quantified using Mander’s colocalization coefficient (ImageJ software) by analyzing a minimum of 100 cells and presented as a dot plot. Red plot represents the overlapping of Endonuclease G over BG4 while green plot represents the overlapping of BG4 over Endonuclease G. (C) Schematic showing the pull-down assay used for evaluation of binding of Endonuclease G present in the rat testicular mitochondrial extracts to the mitochondrial genome. Bound regions were pulled out using anti-Endonuclease G and protein A/G beads. Regions of interest were detected by either semi-quantitative PCR or real-time PCR using appropriate primers. (D) Agarose gel profile showing the amplification through semi-quantitative PCR of Input DNA (upper panel) and Endonuclease G pull down DNA (lower panel). Primers specific to 5 G-quadruplex forming regions (GR1-GR5) and 10 random regions (CR1-CR10) were also used for the amplification. (E) Real-time PCR of 5 G-quadruplex forming regions (blue) and 10 random regions (green) following pull-down assay. Input DNA served as template control. Antibody control served as a negative control. Error bar represents three independent biological repeats. (F) Evaluation of binding of Endonuclease G to different regions of the mitochondrial genome within cells by mito IP. Cells were crosslinked and then mitochondria were isolated. Endonuclease G bound DNA was obtained and was amplified for different regions of mitochondria, as explained in panel E. Graph is plotted for the Ct values obtained following real-time PCR as described above. The error bar represents three independent biological repeats. Refer also Figure 9—figure supplement 1, Figure 2.