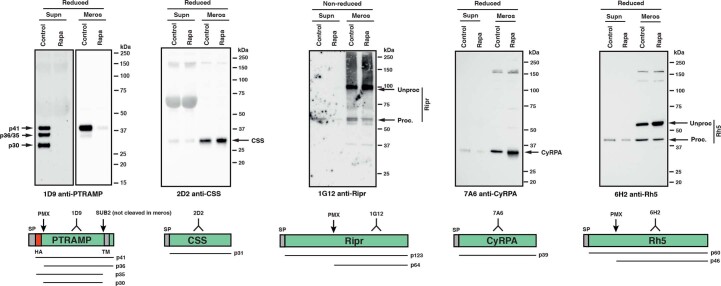
Extended Data Fig. 2. PfCSS, PfRipr, CyRPA and PfRh5 are expressed at normal levels when PfPTRAMP expression is removed by conditional gene knockout.
3D7-PTRAMPiKO was grown without (Control) and with rapamycin (Rapa) to mature schizonts and merozoites and supernatants prepared and analysed by SDS-PAGE and immunoblots that were probed with mAbs, 1D9 (PfPTRAMP), 2D2 (PfCSS), 1G12 (PfRipr), 7A6 (CyRPA) and 6H2 (PfRh5). At bottom of each panel is structure of the relevant protein with the domain recognised by each mAb with respect to PMX protease processing. The PfPTRAMP protein had a HA tag towards the N-terminus (red). The grey domains correspond to the Signal Sequence (SP). The molecular weight of the processed polypeptides and position within each protein is shown. Supernatants were prepared from purified schizonts placed back in culture and allowed to egress. Merozoites were centrifuged as pellets and prepared for SDS-PAGE analysis.