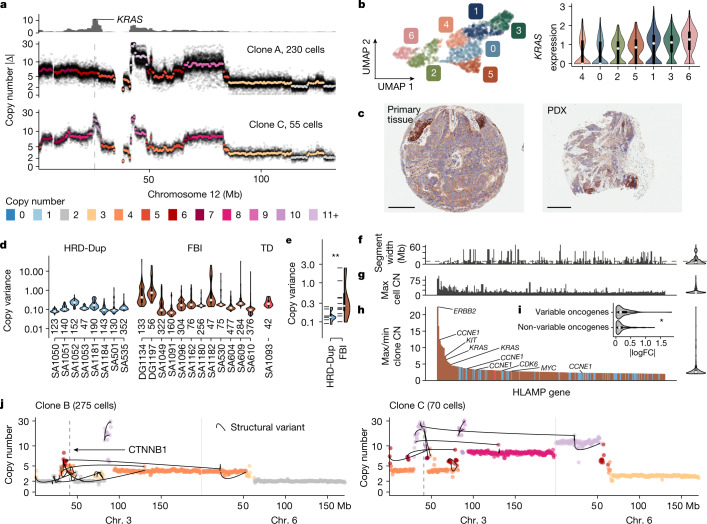
Fig. 4. HLAMP copy number variation.
a, Clone and single-cell whole-genome consensus copy number profiles for chromosome 12 in FBI tumour (SA1049) clones A and C. The top track shows the absolute difference between the copy number of the two clones; the bottom two panels show consensus copy number profiles (coloured points) and all single-cell values (small black points). Cell numbers indicate the number of cells used for consensus copy number evaluation. b, Uniform manifold approximation and projection (UMAP) dimensionality reduction of scRNA-seq data from SA1049 coloured by gene expression cluster and violin plots of gene expression per cluster of KRAS; n = 1,697 cells (per cluster: 0 = 287, 1 = 286, 2 = 276, 3 = 270, 4 = 221, 5 = 200, 6 = 157). c, Immunohistochemistry staining of KRAS protein in primary human tissue (left) and PDX tissue (right). Scale bars, 200 µm (left); 300 µm (right). Images are representative of two cores stained from each PDX tissue. d, Copy number variance across cells for HLAMP bins within each dataset (number of bins shown below each violin). Datasets without HLAMPs not shown. e, Distribution of mean copy variance in eight HRD-Dup cases versus 12 FBI cases. P = 0.0096 (two-sided Wilcoxon test); **P < 0.01. Notches show individual data points. f, Width of genomic segment containing the amplification; dashed red line indicates a width of 10 Mb. g, Maximum cell copy number (CN) per gene. h, Clone maximum/minimum copy number ratio of cancer genes overlapping HLAMP regions. Genes across all cancer datasets with ratio > 2 are shown (n = 296). Colours as in d. For f–h, distributions of values are shown in a violin plot on the right. i, Distribution of the maximum logFC between gene expression clusters in matched scRNA-seq for variable oncogenes (n = 140) versus non-variable oncogenes (n = 159). P = 0.019 (two-sided Wilcoxon test). j, Consensus copy number profiles in two clones in SA1096 overlaid with lines indicating SVs. All box plots indicate the median, first and third quartiles (hinges), and the most extreme data points no farther than 1.5× the IQR from the hinge (whiskers).