FIGURE 2.
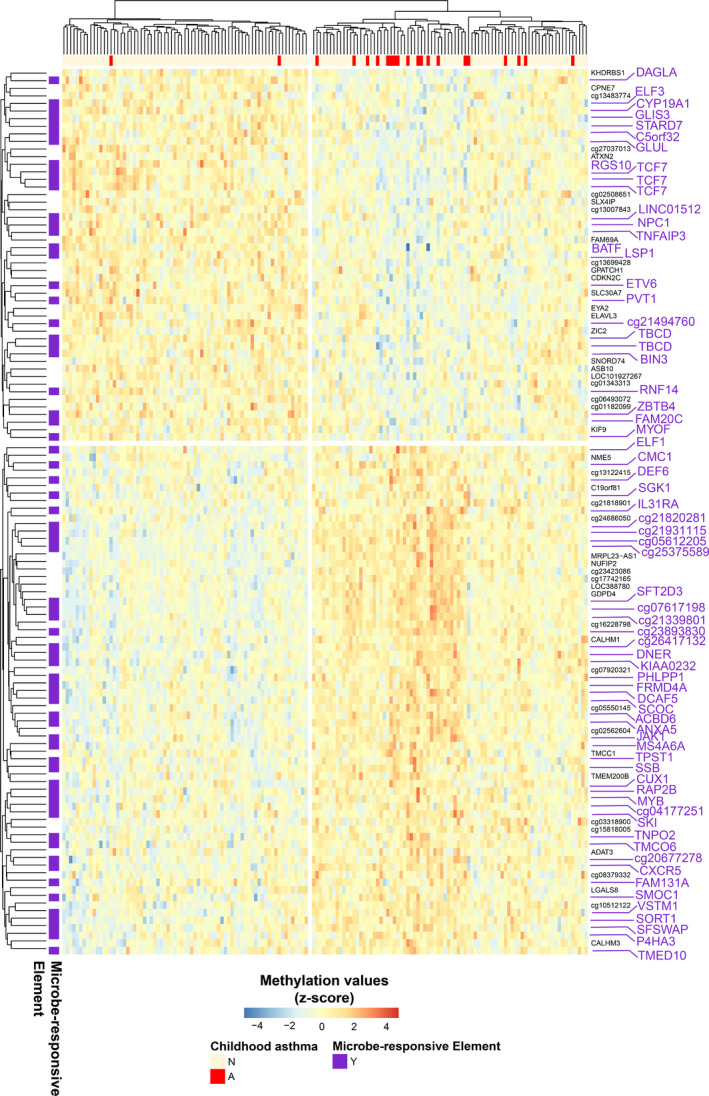
Asthma‐associated differential methylation in the TRQ module co‐localizes with microbe‐responsive elements. Heatmap of TRQ module DMCs (rows) that mapped to promoters/enhancers (as assessed by Roadmap Epigenomics consortium chromatin state annotations) and were significantly (p < .05) associated with asthma. The color bar at the top denotes individual neonates (columns) who did or did not develop asthma during childhood. Microbe‐responsive elements (purple text in the right side legend) were defined as DMCs mapping to genes exhibiting dynamic expression, or putative enhancers exhibiting dynamic changes in chromatin marks (DNaseI/ATAC accessibility, H3K27ac, H3K4me1) in response to microbes or their products. 43 , 44 , 45 , 46 , 47 , 48 , 49 Statistics and annotations for these DMCs are provided in Table S5.
