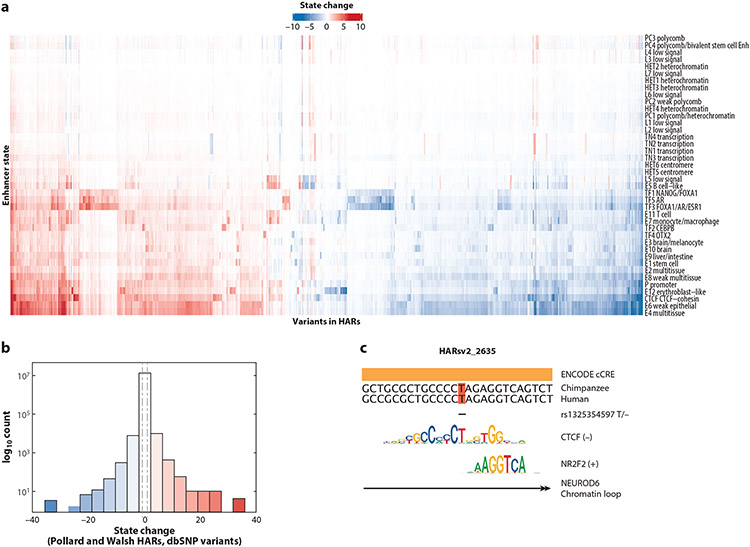
Figure 3.
Deep learning analysis of human variants in human accelerated regions (HARs). All single-nucleotide polymorphisms (SNPs) included on the SNP Database (dbSNP) that overlap with a HAR were scored for their effects on tissue-specific enhancer state predictions using the model Sei (9). This analysis includes all SNPs in all HARs tested in three massively parallel reporter assay (MPRA) studies (22, 66, 71). (a) Increases (red) and decreases (blue) in predicted enhancer state (rows) for all SNPs (columns). (b) Distribution of effects in panel a. Many SNPs in HARs have effect sizes greater than those of known human disease variants [vertical dashed lines represent the median of all SNPs in the Human Gene Mutation Database (HGMD), as reported in Reference 9]. (c) Example of a SNP (rs1325354597) in HARsv2_2635 (22) where the minor allele is predicted to substantially decrease the brain enhancer state and the CTCF state. This variant overlaps an annotated candidate regulatory element (ENCODE cCRE) and motifs of CTCF and NR2F2 as well as other neurological transcription factors (8). The SNP deletes an important nucleotide (T) in the CTCF motif. Consistent with CTCF’s role in loop extrusion, this genomic element has a significant chromatin loop with the promoter of the transcription factor NEUROD6 in cells carrying the major allele (63).