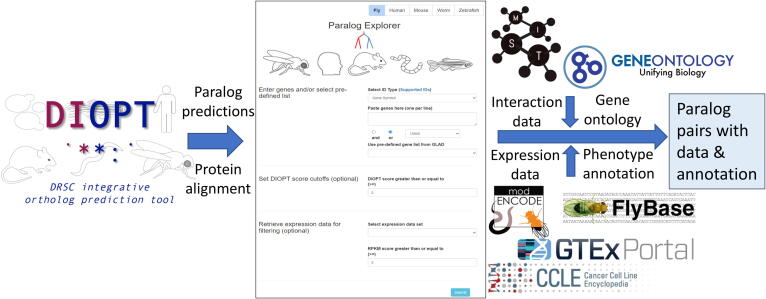
Fig. 1.
Sources for information included in Paralog Explorer Paralog Explorer was built based on the integration of paralog predictions from DIOPT, PPI and genetic-interaction data from MIST, expression data from GTEx and modENCODE as well as gene ontology and phenotype annotation from GO consortium and FlyBase, respectively.