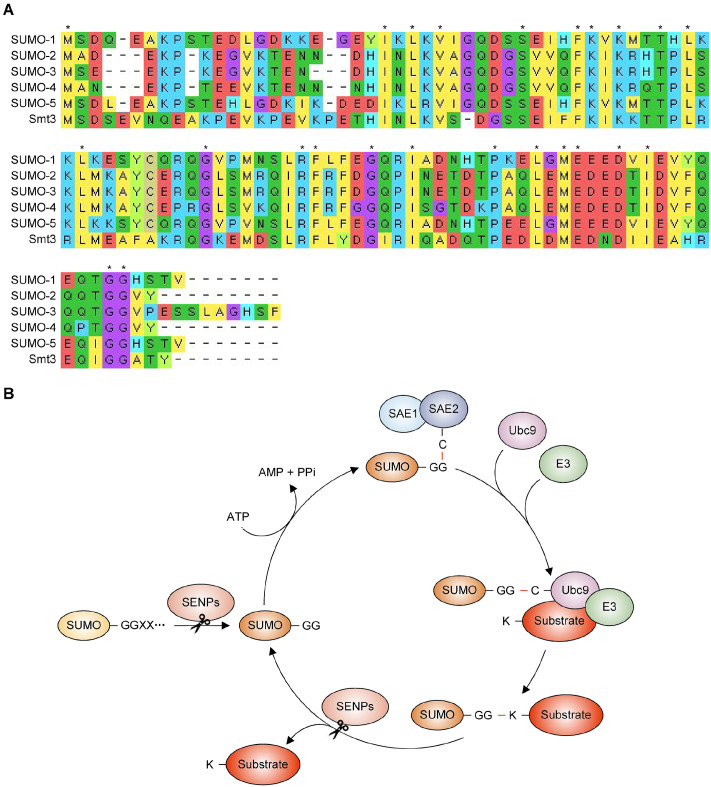
Fig. 1.
Alignment of SUMO protein sequences and SUMO pathway diagram. (A) Sequence alignment of human SUMO-1 through SUMO-5 with yeast SUMO (Smt3) using Molecular Evolutionary Genetic Analysis software (MEGA, https://www.megasoftware.net/). Different letters and colors indicate different amino acids, and asterisks denote amino acids conserved among all SUMO proteins. (B) Diagram of the SUMOylation cycle. The precursor form of SUMO is processed by SENP proteases to create a mature form with a C-terminal Gly-Gly (GG) motif. Mature SUMO is ATP-dependent and activated by heterodimeric E1 SUMO-activating enzymes, SAE1 and SAE2, through a catalytic cysteine (C) residue in SAE2. Next, SUMO is transferred to the C residue of the E2 SUMO-conjugating enzyme (Ubc9), resulting in the conjugation of SUMO to the lysine (K) residues of the substrate protein with the aid of an E3 SUMO ligase. Finally, SENPs can deconjugate SUMO from the substrate or edit SUMO chains, after which SUMO is recycled through the conjugation event.