Fig. 2.
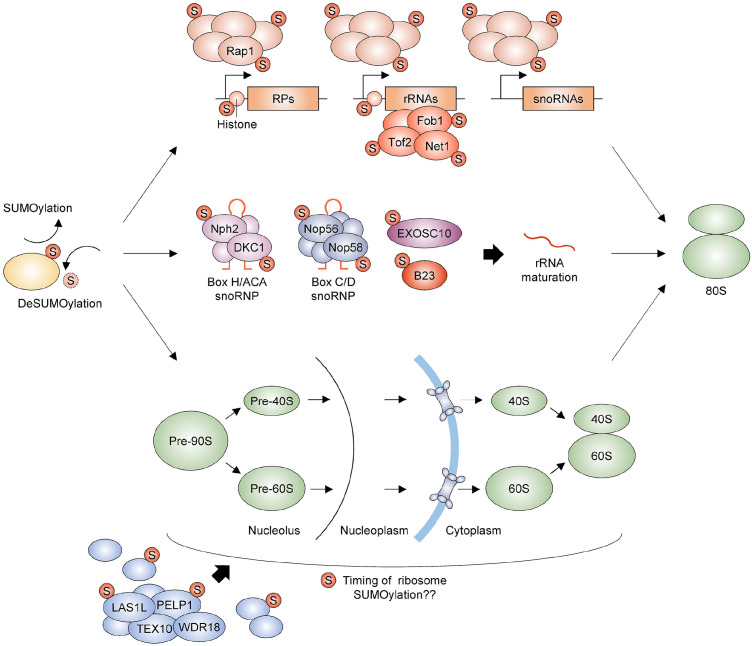
SUMOylation regulates ribosome biogenesis. SUMOylation and deSUMOylation are highly dynamic cellular processes, and their versatile control is essential for ribosome biogenesis. First, several transcription factors, including yeast Rap1, are major substrates of the SUMO pathway. Their SUMOylation regulates the expression of genes encoding ribosomal proteins (RPs) and rRNAs (components of ribosomes) and snoRNAs (required for rRNA maturation). While the association of Net1, Tof2, and Fob1, which are required for rDNA silencing, with rRNA-encoding rDNA locus is regulated by their SUMOylation, SUMOylated histones are substantially located at RP genes and rDNA regions. However, the role of histone SUMOylation in these regions remains unelucidated. Second, the control of SUMOylation of snoRNP proteins, Nph2, DKC1, Nop56, and Nop58, the RNA exosome EXOSC10, and B23/nucleophosmin is required for rRNA maturation. Finally, ribosomal proteins and their assembly factors, including LAS1L and PELP1, are common targets of SUMOylation. However, the timing of SUMOylation required for successful ribosome assembly and export remains unknown. The illustration presents relevant components but not precise physical interactions between proteins or event orders.