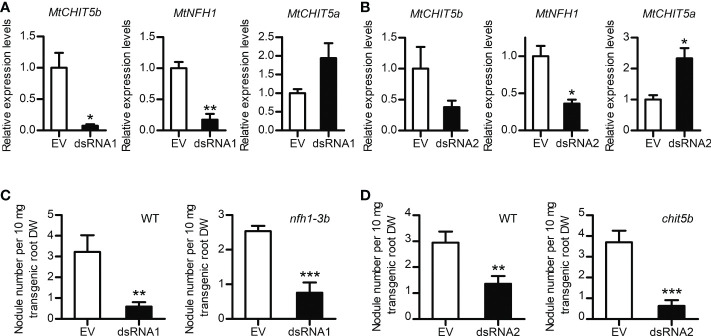
Figure 6.
Nodulation phenotype of M. truncatula roots silenced in MtCHIT5b/MtNFH1 expression by the dsRNA1 and dsRNA2 constructs. (A, B) RT-qPCR analysis of MtCHIT5b, MtNFH1 and MtCHIT5a expression in R108 wild-type roots transformed with the dsRNA1 construct (A). or the dsRNA2 construct (B). Roots transformed with the empty vector (EV) containing RFP alone served as a control. Plants were harvested at 17 days post A. rhizogenes-mediated transformation and co-expression of RFP allowed identification of transgenic roots. Data indicate means ± SE of normalized values (n = 3; mean expression value of the empty vector control set to one) (Student’s t-test; *, P< 0.05; **, P< 0.01). (C, D) Nodule formation on roots of R108 wild-type (WT), nfh1-3b or chit5b mutant plants transformed with the dsRNA1 construct (C) or the dsRNA2 construct (D). Roots transformed with the EV served as a control. Plants with red fluorescent roots were inoculated with S. meliloti Rm41. At the time of harvest (4 wpi), the nodule frequency per root dry weight (DW) was determined for each plant. Data indicate means ± SE (Student’s t test; **, P< 0.01; ***, P< 0.001). Data shown in panel (C) were obtained from 16 transformed WT roots (EV, n = 7; dsRNA1, n = 9) and from 20 transformed nfh1-3b mutant roots (EV, n = 9; dsRNA1, n = 11). Data shown in panel D were obtained from 17 transformed WT roots (EV, n = 9; dsRNA2, n = 8) and 22 transformed chit5b mutant roots (EV, n = 10; dsRNA2, n = 12).