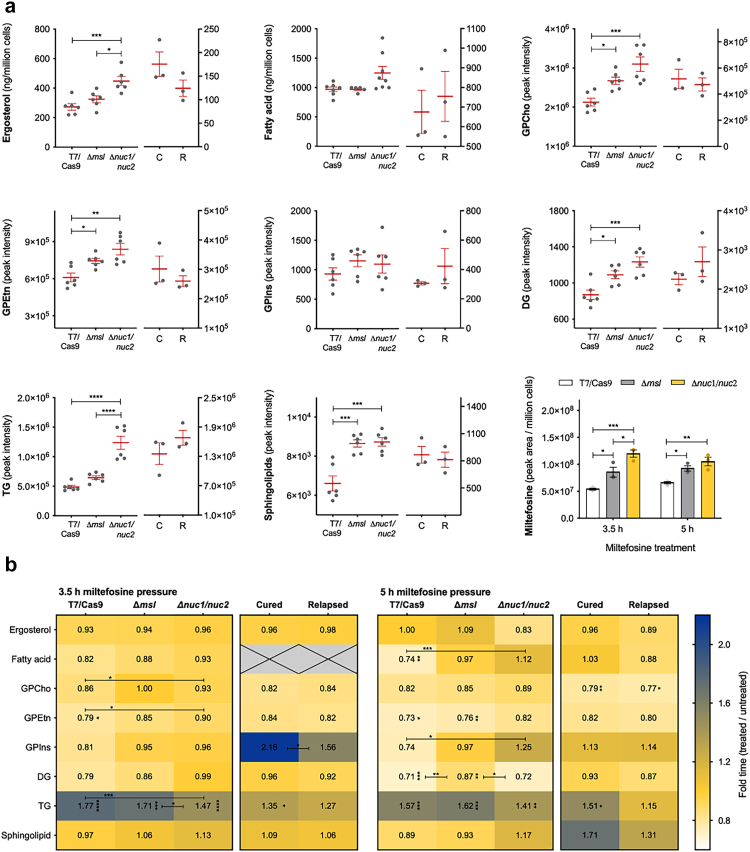
Fig. 5.
Lipidomic analysis of L. infantum. The lipidomic analysis were carried out with parental T7/Cas9, Δmsl and Δnuc1/nuc2, and 6 L. infantum isolates (3 from cured (C)/MSL+/sensitive in vitro and 3 from relapsed (R)/MSL−/resistant in vitro groups) in logarithmic promastigote stage. (a) Lipid content baseline of the L. infantum cell lines and clinical isolates and miltefosine accumulation in the parasite. Each grey dot represents an independent experiment (n = 6) for the isogenic cell lines or the mean of an individual L. infantum isolate from three independent experiments. Miltefosine level was measured after 3.5 h and 5 h treatment with 10 μM of the drug. Mean and ±SEM are shown in red. P values were calculated using one-way ANOVA comparing the cell lines; and using unpaired two-tailed Student's t-tests comparing the group cured with relapsed. (b) Heat map showing the effect of 10 μM of miltefosine on the parasite lipid content. P values were calculated using unpaired two-tailed Student's t-tests comparing: the pairs of untreated with treated, to check if miltefosine treatment is causing a significant perturbation in lipid content (asterisk next to the ratio value in each cell); and the lipid variation caused per miltefosine treatment between the groups of cell lines or clinical isolates, to check if some group is significant more susceptible/resistant to lipid perturbation caused by miltefosine (asterisk over the bar). GPCho, glycerophosphatidylcholines; GPEtn, glycerophosphatidylethanolamines; GPIns, glycerophosphatidylinositols; DG, diacylglycerol; TG, triacylglycerol. ∗ p-value <0.05; ∗∗ p-value <0.01; ∗∗∗ p-value < 0.001; ∗∗∗∗ p-value < 0.0001.