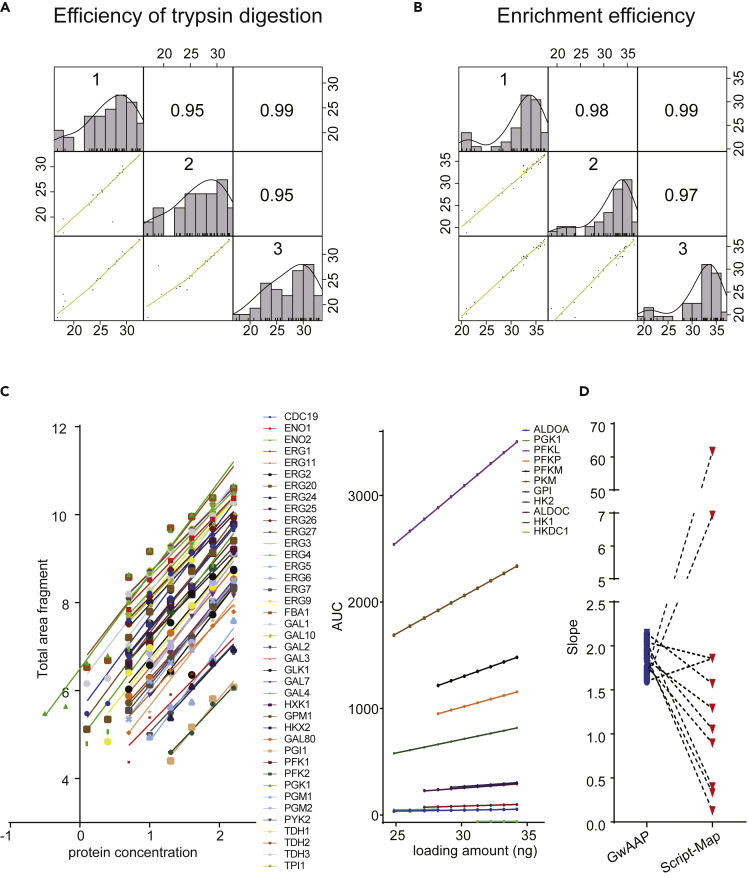
Figure 3.
The reproducibility and linearity of GwAAP
(A) Pearson correlation coefficients within the three independent experiments of trypsin digestion (top right). The samples were digested by trypsin and detected directly by MS to assess the enzymatic efficiency. The 40 protein abundances were log2-transformed in each pairwise comparison, which was plotted as a scatterplot in the bottom left. The green lines represented the least-squares best fit for each pairwise comparison.
(B) Pearson correlation coefficients within the three independent experiments of peptide enrichment (top right). The samples were digested by trypsin, enriched by an HA antibody and detected by MS to assess the enrichment efficiency. The 40 protein abundances were log2-transformed in each pairwise comparison, which was plotted as a scatterplot in the bottom left. The green lines represented the least-squares best fit for each pairwise comparison.
(C) Comparison of the linearity range of dilution curves with the GwAAP and Script-Map approaches. The dilution curves containing 40 code peptides obtained from PRM analysis with the GwAAP approach (left panel). Total area fragment meant the sum of the area under the curve (AUC) of the 6 most intense fragment ions. In contrast, Script-Map measured the linearity range of 11 out of 40 proteins (right panel), except ALDOB, of which the curve slope was −0.081. The AUC was the peak area for the precursor.
(D) Slope comparison of GwAAP (colored in blue) and Script-Map (colored in red). The linearity and coverage of GwAAP are more consistent than those of Script-Map.