Figure 4.
Construction of yeast strains with HA code sequences knock-in in vivo
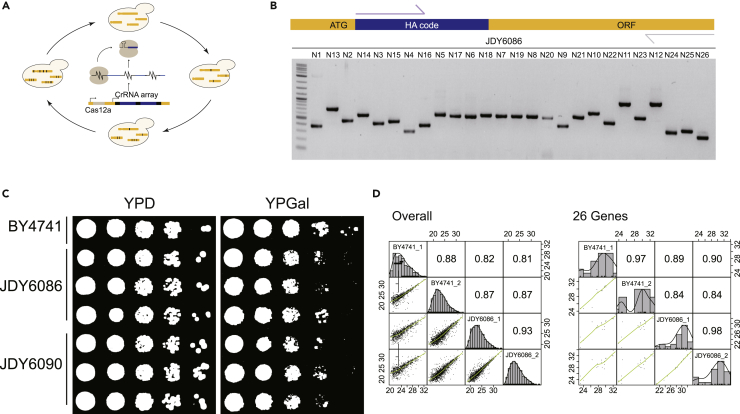
(A) The workflow of a CRISPR-Cas12a-based genome editing system.
(B) PCR analysis of JDY6086, the strain with 26 HA codes knock-in. The forward primer was the same for all the genes and the size of the amplicon for each gene was list in Table S5.
(C) Fitness analysis of BY4741 with or without HA code knock-in on glucose (YPD) and galactose (YPGal) media. All the photos were taken at 48 h JDY6086, strains with 26 HA codes knock-in, 17 glycolytic genes and 9 galactose metabolic genes; JDY6090, strains with 9 HA codes knock-in, 9 galactose metabolic genes.
(D) The correlation of the protein expression levels between BY4741 and JDY6086 cultured in galactose-containing media. The overall proteome (left panel) and the targeted 26 proteins (right panel) were analyzed by DDA mode.