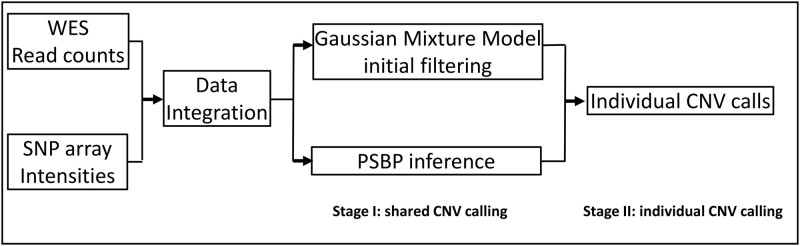
Fig. 1.
Analysis workflow of BMI-CNV. BMI-CNV uses 2 inputs: (1) WES raw read count data from testing and control samples that are computed by using genotyping tools such as SAMtools; and (2) SNP array intensities. WES read counts are normalized to correct exon length, GC-content, and mappability biases. Logarithms of normalized values between testing and pooled control samples are calculated. SNP array intensities are normalized to adjust the genomic waves. The WES and SNP array data are standardized by a robust scaling approach and then integrated. For CNV calling, BMI-CNV carries out a 2-stage framework to generate CNV calls. In stage I, an initial data filtering procedure is coupled with a Bayesian PSBP method to identify shared CNV regions. In stage II, an individual CNV calling procedure is performed to call CNVs in each sample.