Figure 5. Pull‐down of ubiquitylated proteins from phagosomes from WT and RNF115 KO macrophages indicates RNF115 ubiquitylates various proteins involved in immune responses and vesicle trafficking and may affect formation of the VAMP8/STX7 SNARE complex.
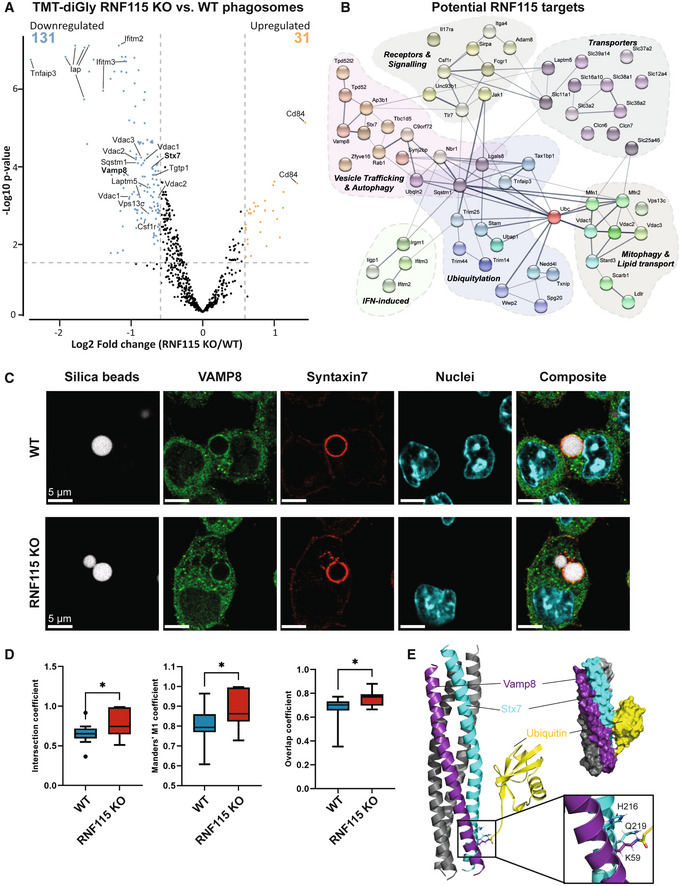
- Volcano plot of proteomics experiment of Gly‐Gly peptides from phagosomal extracts of WT and RNF115 KO macrophages. Selected proteins are highlighted.
- Network representation (from STRING v11) (Szklarczyk et al, 2019) of selected proteins with reduced ubiquitylated peptide abundance in response to loss of RNF115, that is potential phagosomal substrates of RNF115.
- Immunofluorescence micrographs of VAMP8 (green) and Syntaxin‐7 (STX7; red) show strong colocation around 3 μm silica bead phagosomes (white) in BMA macrophages. Nuclei are stained with Dapi. Scale bar is 5 μm. To account for people with red‐green colour‐blindness, we have added a magenta/cyan/yellow version in Appendix Fig S5.
- Colocation of VAMP8 (green) and STX7 (red) on individual phagosomes between WT and RNF115 KO cells represented by intersection, Manders' M1 and overlap coefficients. Data are represented as a box and whisker plot of total values across biological duplicate experiments, where the whiskers represent the minimum to maximum values and the central band indicates the median. Error bars represent SEM of two technical replicates of two biological replicates. *P‐value < 0.05; by unpaired two‐tailed Student's t‐test.
- Structure of VAMP8 and STX7 complex onto which a single ubiquitin molecule on VAMP8 K59 was modelled. Inlay: ubiquitylation of K59 disrupts a hydrogen bond between VAMP8 K59 and STX7 Q219.
