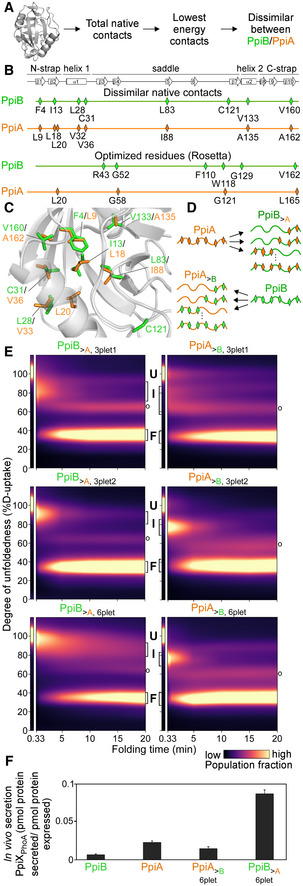
Figure 4. Grafting stable native contacts between PpiB and PpiA interconverted folding behaviours.
-
APipeline for selecting residues that affect folding behaviour using the Frustratometer and 3D structures of PpiB (PDB 2NUL; 1LOP) and PpiA (PDB 1V9T; 1VAI; 1J2A) to test with grafting (details in Dataset EV7).
-
BHighly stabilized, dissimilar native contacts indicated on a linear map with the secondary structural elements on top.
-
CThe side chains of native contact residues (green: PpiB; orange: PpiA) indicated on their 3D structure.
-
DThe native contact grafting scheme between PpiB and PpiA to test their role on folding behaviour.
- E
-
FIn vivo secretion of the indicated PpiX‐PhoA fusion proteins in MC4100 cells carrying SecYprlA4EG. Secretion is expressed as pmol fusion protein secreted from PhoA activity calculations after removing background (uninduced cells) per pmol protein expressed from western blot analysis in 108 cells (Fig EV4E, Dataset EV9). n = 6 (biological triplicates with 3 technical replicates each, s.d.).
Source data are available online for this figure.
