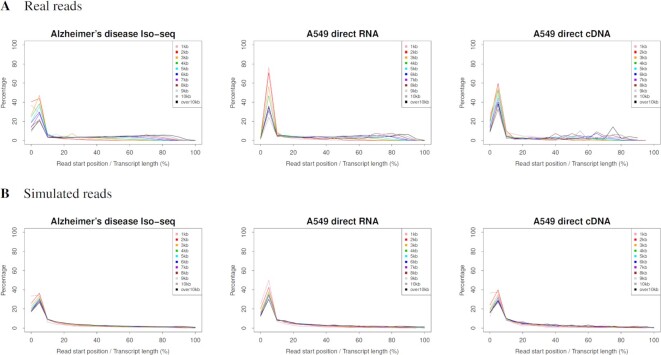
Figure 4.
Read start positions on their template transcripts. Reads were grouped by 1 kb by their template length. Each graph shows the distribution of the read start positions, where colors of plotted lines represent read groups (e.g. 1 kb refers to a read group with their template length of 1–1000 bp). The horizontal axis indicates the position of the read start positions in the total length of their templates, which was calculated by dividing the read start position by the total length of the template; the graph is plotted in 5% increments, with the left edge of the graph showing the percentage of reads starting exactly at the 5’ end of the template. PacBio Iso-seq (CLR read), ONT direct RNA and ONT direct cDNA were simulated using the PBSIM3 quality score models. The Iso-seq (HiFi read) was generated by ccs software as consensus sequences from PBSIM3 outputs. The template transcript from which each read was most likely sequenced and the read start position was obtained from alignments between the reads and their reference transcriptomes.