Figure 5. The RAD51AP1 SUMO-SIM regulatory axis is required for TERRA suppression.
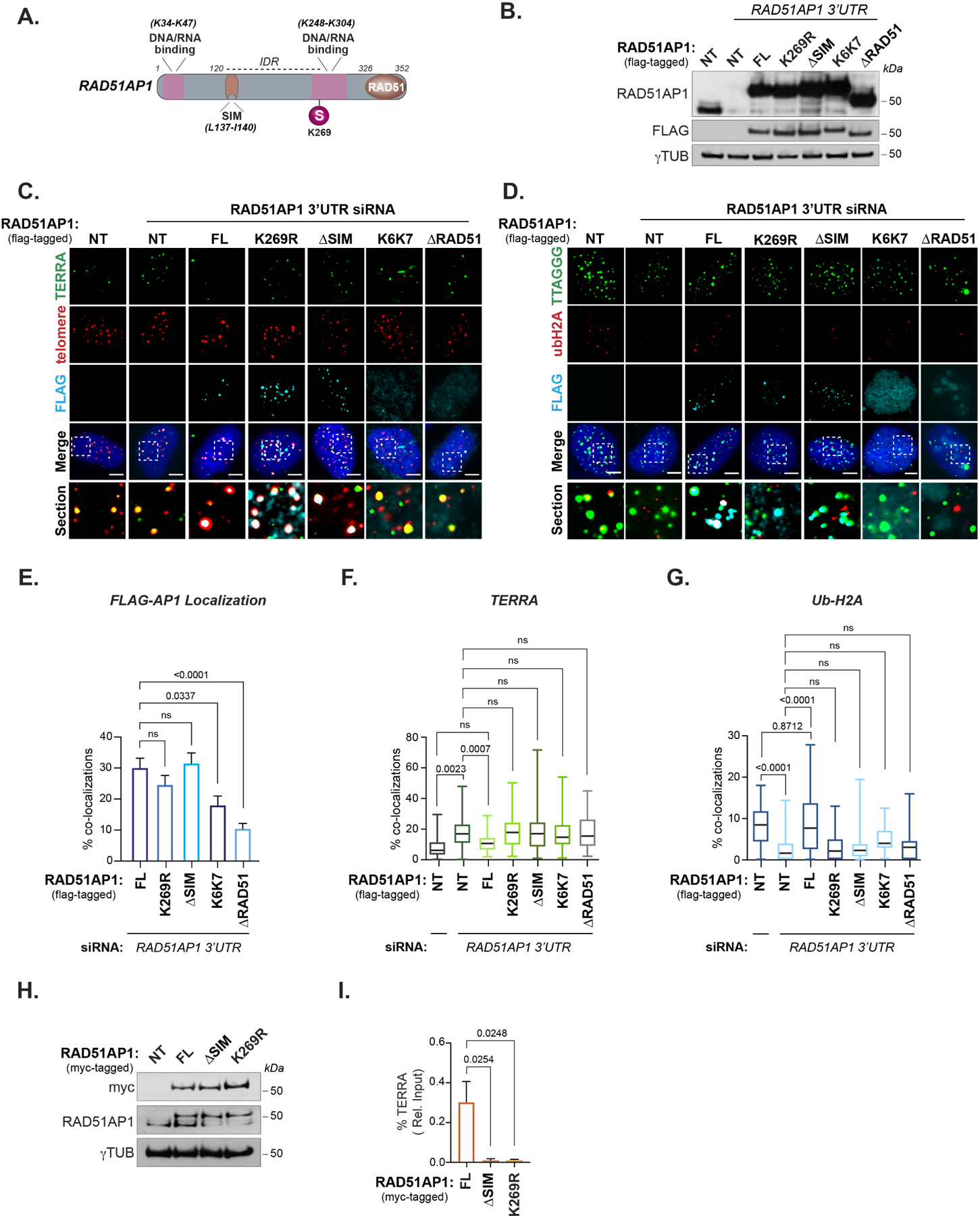
(A) Schematic showing the location of RAD51AP1 functional domains, SUMO modifications and known amino acid substitutions that alter DNA/RNA binding. (B) Western blot of RAD51AP1 knockdown and complementation with wildtype and mutant FLAG-tagged RAD51AP1 proteins. (C) Representative images of TERRA (RNA-FISH) at telomeres (TRF2) after knockdown of RAD51AP1 and complementation with FLAG-tagged RAD51AP1 mutants in U2OS cells synchronized in G2-phase. (D) Representative image of Ub-H2A at telomeres (TRF2) after knockdown of RAD51AP1 and complementation with FLAG-tagged RAD51AP1 mutants in U2OS cells synchronized in G2. (E) Quantification of full-length (FL) and mutant FLAG-tagged RAD51AP1 localization at telomeres in U2OS cells depleted of endogenous RAD51AP1. (F) Quantification of TERRA localization and (G) Ub-H2A accumulation at telomeres. Box and whiskers plots in F and G show the interquartile and min-max ranges. The median is represented by the horizontal black line. Data from triplicate independent experiments are shown. (H) Western blot of U2OS cells stably expressing myc-tagged RAD51AP1. (I) Quantification of TERRA recovered from U2OS cells expressing myc-tagged wildtype, K269R and ΔSIM RAD51AP1. Data represent mean ± SEM, n=3 biological replicates. p values are indicated and generated by One way ANOVA. All scale bars, 5μm.
