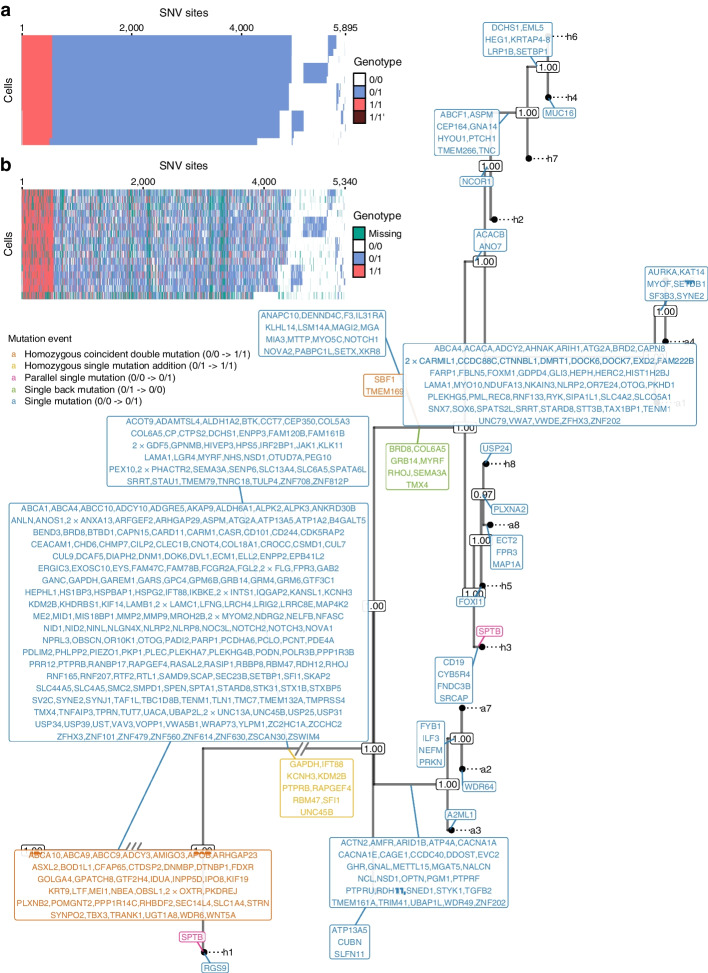
Fig. 4.
Results of phylogenetic inference and variant calling for TNBC16 [42] dataset. Shown is SIEVE’s maximum clade credibility tree. Two exceptionally long branches are folded with the number of slashes proportional to the branch lengths. Tumour cell names are annotated to the leaves of the tree. The numbers at each node represent the posterior probabilities (threshold ). At each branch, genes with non-synonymous mutations are depicted in different colours, representing various types of mutation events. a, b Variant calling heatmap for SIEVE (a) and Monovar (b). Listed in the legend are the categories of predicted genotypes by each method. Cells in the row are in the same order as that of leaves in the phylogenetic tree