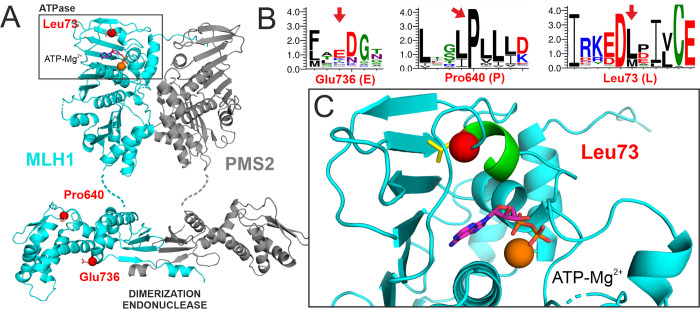
Fig 4. Structural positions of affected residues.
A. The affected residues are shown as red balls in the structural model of the MLH1-PMS2 heterodimer (MLH1 shown in cyan, PMS2 in grey). The dimer comprises N-terminal ATPase domains (top, the bound ATP is shown in stick representation) in which the L73 residue is located. In the C-terminal domains, which confer dimerization and harbor the endonuclease function, the P640 and E736 residues are located. B. Weblogo presentation of sequence conservation of eukaryotic MLH1 in proximity of the three analyzed residues. These residues are marked by red arrows. Letter size corresponds to the degree of conservation. C. Detailed view of leucine 73 (red ball, side chain shown in yellow sticks) within the N-terminal ATPase pocket (area boxed in A). L73 is located within an α-helix (green) that contributes to formation of the nucleotide binding site. It engages in hydrophobic interactions with the extensive β-sheet forming the back of the ATPase domain.