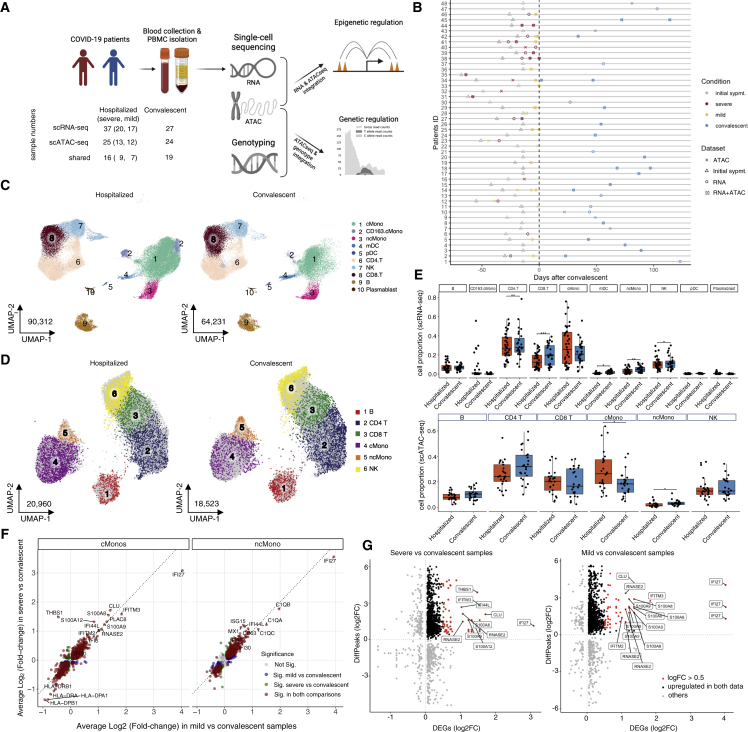
Figure 1.
Study overview and single-cell multi-omics
(A) Workflow of the study. Sample numbers in each data layer and disease condition are indicated.
(B) Schematic overview of all patients enrolled in the study. Sampling dataset, disease conditions, and convalescent days are indicated.
(C and D) UMAP showing the cell distribution of hospitalized and convalescent conditions in scRNA-seq (C) and in scATAC-seq (D); see also Figure S3 and Table S2 for annotation details.
(E) Boxplots showing cell proportion of hospitalized and convalescent samples in main cell types of scRNA-seq and scATAC-seq.
(F) Scatterplots showing the log-fold-change (log2FC) of DE-Gs identified in monocytes between the comparison of severe versus convalescent and the comparison of mild versus convalescent. More details on DE-Gs can be found in Figure S4 and Table S3.
(G) Scatterplots showing the log2FC of DE-Gs and log2FC of differentially accessible peaks (DAPs) identified in classical monocytes between comparison of severe versus convalescent and comparison of mild versus convalescent. Details of the matched DE-Gs and DAPs can be found in Table S4.