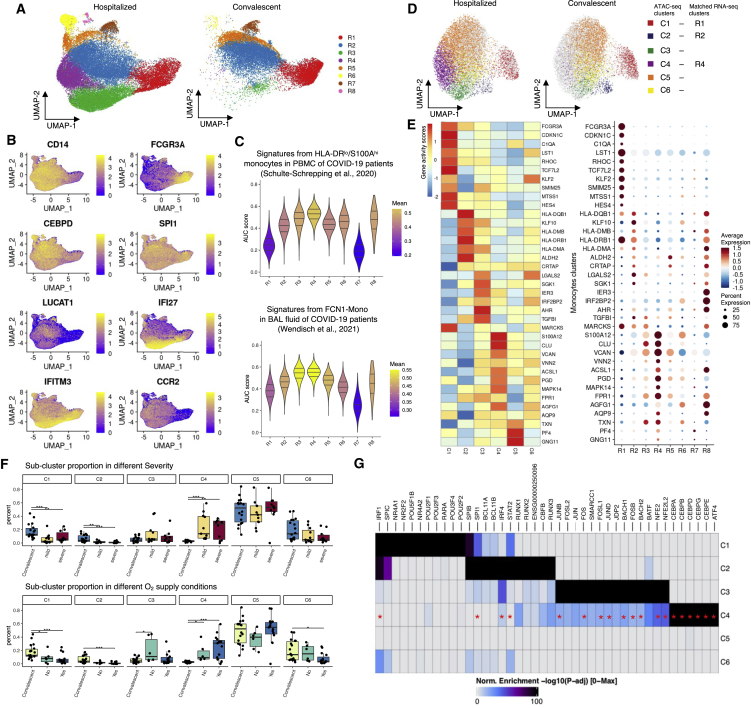
Figure 3.
RNA and ATAC profiles in monocyte sub-clusters in hospitalized and convalescent COVID-19 patients
(A) UMAP showing the cell distribution of hospitalized and convalescent conditions in monocyte sub-clusters of scRNA-seq.
(B) Expression of marker genes in monocyte sub-clusters. See also Table S6 for all the markers.
(C) Violin plots showing the AUCell-based gene signature scores for each sub-cluster from HLA-DRloS100Ahi monocytes in PBMCs (Schulte-Schrepping et al.4) and infiltrating monocytes (FCN1-Mono) in bronchoalveolar lavage (BAL) fluid (Wendisch et al.25).
(D) UMAP showing the monocyte sub-clusters of scATAC-seq.
(E) Dot plots and heatmap showing the expression and imputed activity scores of shared marker genes identified in monocyte sub-clusters of scRNA-seq and scATAC-seq, respectively.
(F) Boxplots showing the cell proportion of severe, mild, and convalescent patients, as well as oxygen supply needed, not needed, and convalescent patients in each monocytes sub-cluster of scRNA-seq. See the cell distributions in Figure S8.
(G) Heatmap showing the significantly enriched TF motifs in the open chromatin peaks of each monocytes sub-cluster; TFs that were also enriched as regulon in R4 cluster cells by SCENIC (single-cell regulatory network inference and clustering) are marked with red asterisks.
See also Figures S6–S8 and Table S7.