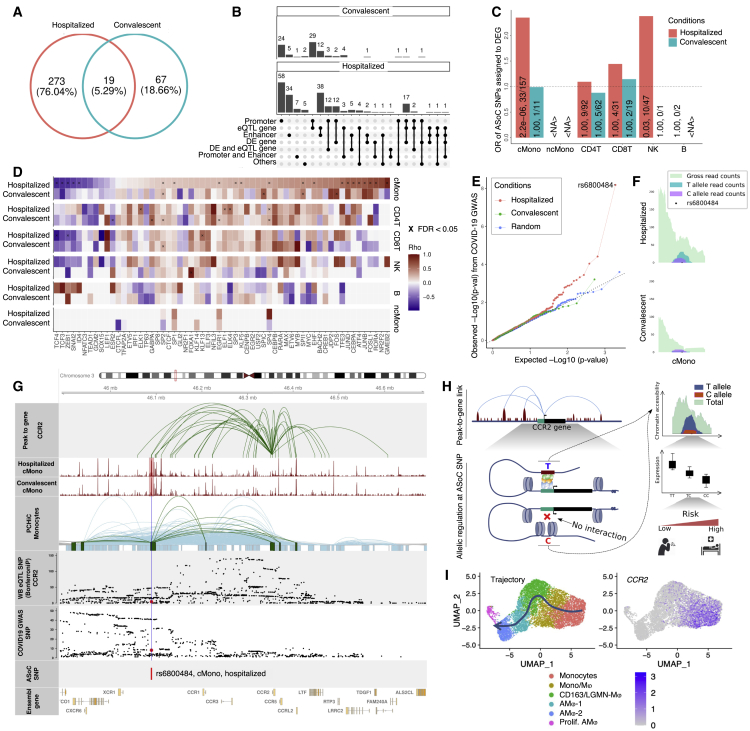
Figure 5.
ASoC analysis reveals the epigenetic effect of COVID-19 GWAS variants
(A) Venn diagram of ASoC SNPs identified in six cell types from hospitalized and convalescent participants. ASoC SNPs were merged per disease condition. ASoC SNPs identified in more than one cell type were counted once.
(B) Upset plot showing functional annotation of identified ASoC SNPs. Regulatory element annotations were determined based on 25-state models from the Roadmap Epigenomics Project. ASoC SNPs were assigned to eQTL genes and DE genes based on significant variant-gene pairs (GTEx V8) and positions (25 kbp up/downstream of ASoC SNPs), respectively. Numbers at the top of each bar indicate the exact number of ASoC SNPs belonging to the annotation or the gene group.
(C) Bar plot showing the enrichment of ASoCs assigned to our DE-Gs. The x axis represents cell types, and the y axis represents odds ratio that ASoCs are assigned to the DE-Gs (i.e., ASoC SNP is located in the promoter of the DEG). Color indicates disease conditions: red for hospitalized COVID-19 and blue for convalescent COVID-19. The numbers on the bar are FDR-adjusted p values and number of ASoC SNPs assigned to DEG out of the number of ASoC SNPs identified for the cell type.
(D) Heatmap of correlations between allelic imbalance and TF motif disruption. For each ASoC SNP, the allelic imbalance was represented by log2(reference read counts/alternative read counts), while the motif disruption was the difference between altScore and refScore by motifbreakR. Colors of the heatmap are Spearman’s rho, and multiplication symbols (×) indicate significant correlations (FDR-adjusted p < 0.05).
(E) Q-Q plot of COVID-19 GWAS p values for identified ASoC SNPs. The y axis represents observed GWAS p values (converted by −log10) of ASoC SNPs in cMono of hospitalized (red), convalescent COVID-19 (green) participants, and random selected SNPs (blue) with matched minor allele frequency.
(F) Allelic reads depth of ASoC SNP rs6800484 at the COVID-19-related CCR locus.
(G) Integration of gene-to-peak link, single-cell ATAC-seq, promoter capture Hi-C, eQTL SNPs, and COVID-19 GWAS SNPs around ASoC SNP rs6800484.
(H) Schematic plot showing the potential epigenetic and genetic regulating program at CCR2 locus under COVID-19 scenario.
(I) CCR2 expression in the differentiation trajectory of monocytes and macrophages of BAL fluid samples of COVID-19 patients (Wendisch et al.25).
<NA>, no valid estimation available. See also Figure S10.