Abstract
Background
Staphylococcus aureus and Streptococcus agalactiae are the main cause of clinical mastitis in dairy cattle in Argentina, whereas coagulase-negative staphylococci (CNS) and environmental streptococci are the main cause of subclinical mastitis. Bacteria isolated from infected animals show increasing antimicrobial resistance.
Objectives
This study aims to determine the antimicrobial resistance of staphylococci and streptococci isolated from milk with mastitis, and to genotypically characterize the methicillin-resistant (MR) staphylococci.
Methods
Isolation was performed on blood agar and identification was based on biochemical reactions. Antimicrobial susceptibility was according to the Clinical and Laboratory Standards Institute guidelines. The antimicrobial resistance genes, SCCmec type and spa type were detected by the polymerase chain reaction method.
Results
We isolated a total of 185 staphylococci and 28 streptococci from 148 milk samples. Among the staphylococcal isolates, 154 were identified as CNS and 31 as S. aureus. Among the 154 CNS, 24.6% (n = 38) were resistant to penicillin, 14.9% (n = 23) to erythromycin, 17.5% (n = 27) to clindamycin, 6.5% (n = 10) to cefoxitin and oxacillin. Among the S. aureus isolates, 16.1% (n = 5) were resistant to penicillin, 3.2% (n = 1) to cefoxitin and oxacillin (MRSA). Six MR isolates (5 CNS and 1 MRSA) were positive to the mecA gene, and presented the SCCmec IVa. The MRSA strain presented the sequence type 83 and the spa type 002. Among the 28 streptococcal isolates, 14.3% (n = 4) were resistant to penicillin, 10.7% (n = 3) to erythromycin and 14.3% (n = 4) to clindamycin.
Conclusions
The present findings of this study indicate a development of antimicrobial resistance in main bacteria isolated from cows with mastitis in Argentina.
Keywords: Bovine mastitis, antimicrobial resistance, staphylococci, streptococci, molecular typing
INTRODUCTION
Bovine mastitis, which is the inflammation of the mammary gland tissue, usually has an infectious etiology. The cost of the cow's treatment and the decrease in milk production makes mastitis the most costly disease to the dairy industry [1]. The main microorganisms causing bovine mastitis in Argentina and many other countries around the world are Gram-positive cocci, mainly streptococci and staphylococci [2]. Among streptococci, Streptococcus agalactiae is one of the main causes of subclinical mastitis in dairy cattle, despite its antimicrobial susceptibility [3], while Streptococcus dysgalactiae and Streptococcus uberis are environmental pathogens [4]. Among staphylococci, Staphylococcus aureus is the most important pathogen in bovine clinical mastitis, while coagulase-negative staphylococci (CNS) are more frequent in subclinical mastitis cases [5]. These microorganisms currently show increasing antimicrobial resistance and are considered reservoirs of resistance genes [1]. Antimicrobial resistance has become a concern for many reasons, including therapeutic failure, economic losses and public health repercussions. In addition, resistant pathogens may spread and become a serious problem in healthcare institutions and communities [6]. Thus, to select an appropriate antimicrobial therapy, it is important to correctly identify the pathogens involved [7]. Furthermore, to determine strategies to prevent the development of antimicrobial resistance and minimize the risk of spreading, it is important to consider the individual susceptibility of the microorganism, the pharmacokinetics of the drug, the time of action in the site of infection, and the animal toxicity [8].
The antibiotics frequently used in intramammary infusion therapy are β-lactam antibiotics. However, methicillin-resistant (MR) staphylococci are resistant to all β-lactam antibiotics, because these bacteria acquire a penicillin-binding protein called PBP2a, which has low affinity for these drugs. This low-affinity protein is encoded by the mecA/C genes located on a mobile genetic element called staphylococcal chromosomal cassette (SCCmec). SCCmec has been classified into 13 different types (types I-XIII) and subtypes because of differences in the structural organization and genetic content [9]. Originally, mecALGA251 was described as an allele of mecA and has been identified in MR Staphylococcus aureus (MRSA) from both humans and animals. This mecA homolog has been renamed as mecC because it shares 70% nucleotide identity with mecA [10]. In South America, we detected the first mecC gene in a CNS isolated from bovine mastitis [11].
Molecular typing methods are used to find out the clonal relatedness among isolates to monitor and control outbreaks and to determine routes of transmission. Among these, the method with the most discriminatory power is Whole-genome sequencing (WGS) followed by Next-generation sequencing (NGS), pulsed-field gel electrophoresis (PFGE), spa typing and multilocus sequence typing (MLST) [12].
The aim of this study was to identify and detect antimicrobial susceptibility in staphylococci and streptococci isolated from milk samples from mastitic cows in several herds in Argentina, and to determine the occurrence and epidemiology of MR strains based on their antimicrobial resistance patterns and molecular typing.
MATERIALS AND METHODS
Isolation and identification of staphylococci and streptococci
A total of 148 milk samples were collected from cows with subclinical and clinical mastitis between June 2016 and December 2017 in the dairy region of Buenos Aires and Córdoba, Argentina. Briefly, 10 μl of each milk sample was spread over a Columbia blood agar plate and incubated at 37°C for 24-48 h. S. aureus isolates were identified by Gram-staining, catalase, fermentation of glucose, coagulase, fermentation of mannitol, maltose and trehalose, and Voges-Proskauer. To identify the CNS group, we also evaluated oxidase, beta galactosidase and resistance to novobiocin. Streptococcal isolates were identified by catalase, pattern of hemolysis, growth in broth with 4% and 6.5% of sodium chloride, hydrolysis of esculin, the Pyroglutamic acid β-naphthylamide reaction (PYR) and the CAMP test.
Antimicrobial susceptibility testing
Staphylococcal isolates were tested for antimicrobial susceptibility using five antibiotics: penicillin, clindamycin, erythromycin, cefoxitin and oxacillin, and for streptococcal isolates, three antibiotics were tested: penicillin, clindamycin and erythromycin, using the disk diffusion method by Kirby-Bauer and categorized in accordance with the Clinical and Laboratory Standards Institute criteria [13]. The MICs for erythromycin, clindamycin and penicillin were determined by the agar dilution method [13].
Genotyping of MR-CNS and MRSA
The DNA was extracted using the Wizard Genomic DNA purification kit (Promega, USA). The polymerase chain reaction (PCR) mixture for each reaction was: 200 ng of DNA, 0.2 mM dNTP, 1.5 mM MgCl2, 0.5 µM of each primer, 5 µL of buffer 10X, 1.25 U Taq DNA polymerase (BioLabs) and distilled water to a final volume of 25 µL. The cycles and conditions used were described by Vannuffel et al. [14] and Cuny and Witte [15]. SCCmec types were determined by PCR [16,17]. The MRSA identified was analyzed as described by Enright et al. [18]. Spa typing was performed as described by Harmsen et al. [19] and Hallin et al. [20].
Statistical analysis
To compare the results obtained with the data from previous studies carried out by this working group, χ2 statistic and Fisher's exact test were performed using the SPSS software (SPSS Inc., USA) and the values of p < 0.05 were considered statistically significant.
RESULTS
Identification
A total of 185 staphylococci and 28 streptococci were isolated from 148 milk samples from eight farms. Among staphylococcal isolates, 154 were identified as CNS (59.8% S. epidermidis, 20.2% S. simulans, 1.9% S. sciuri and 17.6% S. saprophyticus) and 31 as S. aureus. Among streptococci, eleven were identified as Streptococcus dysgalactiae, four as Streptococcus uberis, six as belonging to the Streptococcus bovis group and seven could not be identified by biochemical tests.
Antimicrobial resistance
Among the 154 CNS, 24.6% (n = 38) were resistant to penicillin, 14.9% (n = 23) to erythromycin, 17.5% (n = 27) to clindamycin, and 6.5% (n = 10) to cefoxitin and oxacillin (MR). Among the 31 S. aureus isolates, 16.1% (n = 5) were resistant to penicillin and 3.2% (n = 1) to cefoxitin and oxacillin (MRSA). Regarding streptococci, 14.3% were resistant to penicillin and clindamycin and 10.7% were resistant to erythromycin (Table 1).
Table 1. Antimicrobial resistance of staphylococci (n = 185) and streptococci (n = 28) isolated from bovine mastitis.
| Antimicrobials | CNS (n = 154) | S. aureus (n = 31) | Streptococcus spp.(n = 28) |
|---|---|---|---|
| PEN | 38 (24.6) | 5 (16.1) | 4 (14.3) |
| FOX/OXA | 10 (6.5) | 1 (3.2) | ND |
| ERY | 23 (14.9) | 0 | 3 (10.7) |
| CLI | 27 (17.5) | 0 | 4 (14.3) |
Values are presented as number (%).
CNS, coagulase-negative staphylococci; PEN, penicillin; FOX, cefoxitin; OXA, oxacillin; ERY, erythromycin; CLI, clindamycin; ND, not determined.
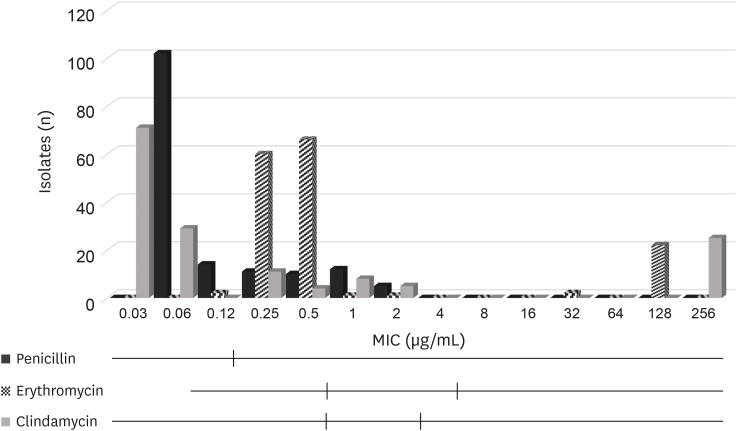
For CNS, the MIC50 and MIC90 values for penicillin were 0.06 µg/mL and 1 µg/mL respectively, those for erythromycin were 0.5 µg/mL and 128 µg/mL respectively, and those for clindamycin were 0.06 µg/mL and 256 µg/mL respectively (Table 2, Fig. 1).
Table 2. MIC values for n = 154 coagulase-negative staphylococci isolated from bovine mastitis.
| Antimicrobials | MIC | Resistance | ||
|---|---|---|---|---|
| 50 | 90 | Number | % | |
| Penicillin | 0.06 | 1 | 38 | 24.6 |
| Erythromycin | 0.5 | 128 | 23 | 14.9 |
| Clindamycin | 0.06 | 256 | 27 | 17.5 |
MIC, minimum inhibitory concentration.
Fig. 1. Distribution of MIC values for n = 154 coagulase-negative staphylococci isolated from bovine mastitis. Black vertical lines indicate the Clinical and Laboratory Standards Institute breakpoints used to classify the isolates as susceptible, intermediate (if available) or resistant.
MIC, minimum inhibitory concentration.
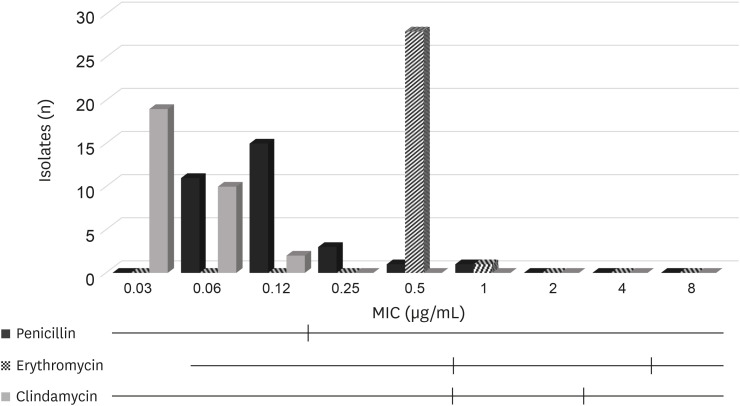
For S. aureus, the MIC50 and MIC90 values for penicillin were both 0.12 µg/mL, those for erythromycin were both 0.5 µg/mL, and those for clindamycin were 0.03 µg/mL and 0.06 µg/mL respectively (Table 3, Fig. 2).
Table 3. MIC values for n = 31 S. aureus isolated from bovine mastitis.
| Antimicrobials | MIC | Resistance | ||
|---|---|---|---|---|
| 50 | 90 | Number | % | |
| Penicillin | 0.12 | 0.12 | 5 | 16 |
| Erythromycin | 0.5 | 0.5 | 0 | 0 |
| Clindamycin | 0.03 | 0.06 | 0 | 0 |
MIC, minimum inhibitory concentration.
Fig. 2. Distribution of MIC values for n = 31 S. aureus isolated from bovine mastitis. Black vertical lines indicate the Clinical and Laboratory Standards Institute breakpoints used to classify the isolates as susceptible, intermediate (if available) or resistant.
MIC, minimum inhibitory concentration.
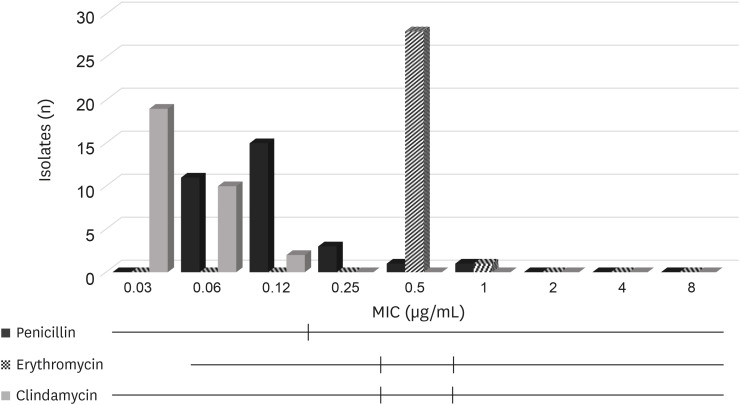
For streptococci, the MIC50 and MIC90 values for penicillin were 0.03 µg/mL and 1 µg/mL respectively, those for erythromycin were 0.12 µg/mL and 0.5 µg/mL respectively and those for clindamycin were 0.06 µg/mL and 4 µg/mL respectively (Table 4, Fig. 3).
Table 4. MIC values for n = 28 streptococci isolated from bovine mastitis.
| Antimicrobials | MIC | Resistance | ||
|---|---|---|---|---|
| 50 | 90 | Number | % | |
| Penicillin | 0.03 | 1 | 4 | 14 |
| Erythromycin | 0.12 | 0.5 | 3 | 11 |
| Clindamycin | 0.06 | 4 | 4 | 14 |
MIC, minimum inhibitory concentration.
Fig. 3. Distribution of MIC values for n = 28 streptococci isolated from bovine mastitis. Black vertical lines indicate the Clinical and Laboratory Standards Institute breakpoints used to classify the isolates as susceptible, intermediate (if available) or resistant.
MIC, minimum inhibitory concentration.
Genotypic characterization
Among MR isolates, five CNS and one MRSA were positive to the gene mecA by PCR, and all of them presented the SCCmec IVa. Methicillin-susceptible isolates were negative for the mecA/C genes. The MRSA strain detected in this study belonged to sequence type (ST) 83 and spa type 002.
DISCUSSION
CNS are the most frequent bacteria isolated from bovine mastitis in Argentina [21] as well as in other countries such as China (73%) and Switzerland (66%) [22,23].
In this study, staphylococci were the most prevalent bacteria isolated, being S. epidermidis and S. simulans the species most commonly found, in accordance with that found in Switzerland (26% and 22% respectively) [23]. However, S. chromogenes was the most prevalent isolate in Brazil (42.5%) [24] and in previous studies in Argentina (46.6%, 44.4% respectively) [21,25].
In this study, S. aureus was found in low percentage (14.5%) as compared with previous studies in our country (60.7%) [26] and other studies carried out in different countries: Colombia (65%) [27] and India (60.87%) [28].
Regarding streptococci, S. dysgalactiae was the most frequent isolate of this genus, in agreement with that reported in India [28]. On the other hand, in Mongol, S. agalactiae was the most prevalent [4], and S. uberis the most prevalent in a previous study in Argentina [29]. The CNS antimicrobial resistance values here found for erythromycin, penicillin and oxacillin were lower than those reported by Raspanti et al. [1] in Argentina (29.2%, 51.6%, and 13.7%, respectively), but higher than those previously reported by us in 2002 (6.5%, 18%, and 4% respectively) [7]. In Poland and China, penicillin resistance has been found to be higher (30% and 55.5% respectively) [30,31]. In Brazil [32], resistance to both penicillin and oxacillin has been found to be higher (79% and 29% respectively), and some reports have shown 100% resistant for penicillin [33], while resistance to erythromycin and clindamycin has been found to be lower (37.5% for both). In Poland, erythromycin resistance has been found to be lower (13.7%) [30].
All S. aureus isolates detected in this study were susceptible to erythromycin and clindamycin, and showed lower resistance to penicillin resistance than in previous years (23.1%) [34]. We reported the first MRSA from milk samples in Argentina [35], which would indicate a change or evolution in antimicrobial resistance. The emergence of MR is due to the acquisition and insertion of the SCCmec into the chromosome. This acquisition of antimicrobial resistance has presented a challenge to the medical world in terms of limits of treatment and control of staphylococcal infections. As S. aureus is known to carry an arsenal of virulence factors, is one of the major causes of hospital and community-acquired infections, resulting in serious consequences [12]. There are reports showing that humans in contact with livestock are at high risk of becoming colonized and infected with livestock-associated MRSA (LA-MRSA). These studies suggest that livestock and other animals may become a permanent reservoir for human MRSA infections [12].
The streptococcal isolates showed higher antimicrobial resistance for the drugs tested than that previously reported in Argentina (0% for penicillin, 1.9% for erythromycin, 1.9% for clindamycin) [36], but lower than that reported in China [4] (95% for penicillin, 66.7% for erythromycin, 65.4% for clindamycin).
Epidemiological cutoff values (ECOFFs) distinguish between organisms with and without phenotypically expressed resistance mechanisms for a bacterial species and a corresponding antibiotic. These two groups are termed “non-wild-type” and “wild-type” respectively. ECOFFs are only used to detect isolates with acquires resistance to an antibiotic. A publicly available database for identifying ECOFFs is available on the EUCAST website (www.eucast.org). Epidemiological cutoffs are microbiological parameters used by EUCAST in the process of clinical breakpoints setting and correspond to the upper MIC/lower inhibition zone size of a wild-type distribution. The upper MIC value of the wild-type distribution separates microorganisms without (wild-type) and with acquired resistance mechanisms (non-wild-type) to the agent in question. The majority of isolates in this study showed a wild-type distribution. In Table 1 we can observe the number of isolates showing a non-wild-type distribution. Acquired resistance mechanisms can be detected by PCR or using modern molecular techniques as WGS, and can be used to validate ECOFFs that have been estimated from phenotypic data [37]. In our study six MR staphylococci were positive to mecA gene. Other resistance genes that can be detected in staphylococcal specie were reported in a previous publication [25]. Several mechanisms of antibiotic resistance can also be detected in streptococci [38].
The SCCmec type IVa detected in MRSA and MR-CNS isolates is frequent in isolates recovered from milk [22,30]. However, in China, the most frequent has been found to be the SCCmecXII [39], and in Italy the SCCmecV [40]. In contrast, the sequence type of the MRSA found in this study is not the most frequently found in milk from mastitic cows [22,41]. In fact, there is not much information of ST83, except that in human cases located in Chile, Poland, Spain and the Gambia reported in the database www.mslt.net.com.
In China, the most prevalent ST found in MRSA are ST71 and ST97 [22] and in Italy the most prevalent are ST152 and ST398 [40]. Interestingly, in Argentina, we have previously reported ST97, ST705, ST746, ST2102 and ST2187 in methicillin-sensitive S. aureus [34]. Regarding spa typing, Asadollahi et al., found that spa types t002 and t008 were the most frequently repeated spa type in 16 countries of different continents [42].
CNS were the most frequent bacteria isolated in this study. The resistance profile for CNS was higher than that found in previous studies in our country, but lower than that found in other countries. S. aureus susceptibility was high and streptococci showed higher resistance than that previously reported in Argentina but lower than that previously reported in other countries. Five MR-CNS were negative for the mecA/C genes, probably because they carry some other resistance mechanism. SCCmecVIa remains the most frequent in mastitic milk. There is no information of this ST in animals. The report of an MRSA isolate from milk from cows with mastitis in Argentina indicates the development of antimicrobial resistance and the necessity for a continuous monitoring for a rational management of drugs in mastitis therapy. Monitoring antimicrobial profiles in animals is important to collect data for trends in antimicrobial resistance phenotypes and genotypes to identify new or emerging resistance profiles.
Footnotes
Funding: This study was supported by the Secretaría de Ciencia y Técnica, Universidad de Buenos Aires, Project 20020170100084BA.
This paper was published with special support from the Korean Society of Veterinary Science.
Conflicts of Interest: The authors declare no conflicts of interest.
- Conceptualization: Gentilini ER, Srednik ME.
- Data curation: Crespi E.
- Formal analysis: Crespi E.
- Funding acquisition: Gentilini ER.
- Investigation: Crespi E, Pereyra AM, Puigdevall T, Rumi MV, Testorelli MF, Gulone L, Srednik ME.
- Methodology: Gentilini ER, Srednik ME.
- Project administration: Gentilini ER, Srednik ME.
- Resources: Caggiano N.
- Software: Crespi E.
- Supervision: Mollerach M, Gentilini ER, Srednik ME.
- Validation: Crespi E.
- Writing - original draft: Crespi E, Srednik ME.
- Writing - review & editing: Crespi E, Mollerach M, Gentilini ER, Srednik ME.
- Visualization: Crespi E, Srednik ME.
References
- 1.Raspanti CG, Bonetto CC, Vissio C, Pellegrino MS, Reinoso EB, Dieser SA, et al. Prevalence and antibiotic susceptibility of coagulase-negative Staphylococcus species from bovine subclinical mastitis in dairy herds in the central region of Argentina. Rev Argent Microbiol. 2016;48(1):50–56. doi: 10.1016/j.ram.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Tenhagen BA, Köster G, Wallmann J, Heuwieser W. Prevalence of mastitis pathogens and their resistance against antimicrobial agents in dairy cows in Brandenburg, Germany. J Dairy Sci. 2006;89(7):2542–2551. doi: 10.3168/jds.S0022-0302(06)72330-X. [DOI] [PubMed] [Google Scholar]
- 3.Keefe GP. Streptococcus agalactiae mastitis: a review. Can Vet J. 1997;38(7):429–437. [PMC free article] [PubMed] [Google Scholar]
- 4.Ding Y, Zhao J, He X, Li M, Guan H, Zhang Z, et al. Antimicrobial resistance and virulence-related genes of Streptococcus obtained from dairy cows with mastitis in Inner Mongolia, China. Pharm Biol. 2016;54(1):162–167. doi: 10.3109/13880209.2015.1025290. [DOI] [PubMed] [Google Scholar]
- 5.Tenhagen BA, Hansen I, Reinecke A, Heuwieser W. Prevalence of pathogens in milk samples of dairy cows with clinical mastitis and in heifers at first parturition. J Dairy Res. 2009;76(2):179–187. doi: 10.1017/S0022029908003786. [DOI] [PubMed] [Google Scholar]
- 6.Tenover FC. Mechanisms of antimicrobial resistance in bacteria. Am J Infect Control. 2006;34(5) Suppl 1:S3–SS10. doi: 10.1016/j.ajic.2006.05.219. [DOI] [PubMed] [Google Scholar]
- 7.Gentilini E, Denamiel G, Betancor A, Rebuelto M, Rodriguez Fermepin M, De Torrest RA. Antimicrobial susceptibility of coagulase-negative staphylococci isolated from bovine mastitis in Argentina. J Dairy Sci. 2002;85(8):1913–1917. doi: 10.3168/jds.s0022-0302(02)74267-7. [DOI] [PubMed] [Google Scholar]
- 8.McDougall S. Efficacy of two antibiotic treatments in curing clinical and subclinical mastitis in lactating dairy cows. N Z Vet J. 1998;46(6):226–232. doi: 10.1080/00480169.1998.36094. [DOI] [PubMed] [Google Scholar]
- 9.Baig S, Johannesen TB, Overballe-Petersen S, Larsen J, Larsen AR, Stegger M. Novel SCCmec type XIII (9A) identified in an ST152 methicillin-resistant Staphylococcus aureus . Infect Genet Evol. 2018;61:74–76. doi: 10.1016/j.meegid.2018.03.013. [DOI] [PubMed] [Google Scholar]
- 10.Petersen A, Stegger M, Heltberg O, Christensen J, Zeuthen A, Knudsen LK, et al. Epidemiology of methicillin-resistant Staphylococcus aureus carrying the novel mecC gene in Denmark corroborates a zoonotic reservoir with transmission to humans. Clin Microbiol Infect. 2013;19(1):E16–E22. doi: 10.1111/1469-0691.12036. [DOI] [PubMed] [Google Scholar]
- 11.Srednik ME, Archambault M, Jacques M, Gentilini ER. Detection of a mecC-positive Staphylococcus saprophyticus from bovine mastitis in Argentina. J Glob Antimicrob Resist. 2017;10:261–263. doi: 10.1016/j.jgar.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 12.Lakhundi S, Zhang K. Methicillin-Resistant Staphylococcus aureus: Molecular Characterization, Evolution, and Epidemiology. Clin Microbiol Rev. 2018;31(4):1–103. doi: 10.1128/CMR.00020-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clinical and Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals. Approved Standard Fourth Edition and supplement VET01-A4 and VET01-S2 (Replaces M31 A3) Wayne: Clinical Laboratory Standard Institute; 2013. [Google Scholar]
- 14.Vannuffel P, Laterre PF, Bouyer M, Gigi J, Vandercam B, Reynaert M, et al. Rapid and specific molecular identification of methicillin-resistant Staphylococcus aureus in endotracheal aspirates from mechanically ventilated patients. J Clin Microbiol. 1998;36(8):2366–2368. doi: 10.1128/jcm.36.8.2366-2368.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cuny C, Witte W. PCR for the identification of methicillin-resistant Staphylococcus aureus (MRSA) strains using a single primer pair specific for SCCmec elements and the neighbouring chromosome-borne orfX . Clin Microbiol Infect. 2005;11(10):834–837. doi: 10.1111/j.1469-0691.2005.01236.x. [DOI] [PubMed] [Google Scholar]
- 16.Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, Etienne J, et al. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother. 2007;51(1):264–274. doi: 10.1128/AAC.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Milheiriço C, Oliveira DC, de Lencastre H. Multiplex PCR strategy for subtyping the staphylococcal cassette chromosome mec type IV in methicillin-resistant Staphylococcus aureus: ‘SCCmec IV multiplex’. J Antimicrob Chemother. 2007;60(1):42–48. doi: 10.1093/jac/dkm112. [DOI] [PubMed] [Google Scholar]
- 18.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus . J Clin Microbiol. 2000;38(3):1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harmsen D, Claus H, Witte W, Rothgänger J, Claus H, Turnwald D, et al. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 2003;41(12):5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hallin M, Deplano A, Denis O, De Mendonça R, De Ryck R, Struelens MJ. Validation of pulsed-field gel electrophoresis and spa typing for long-term, nationwide epidemiological surveillance studies of Staphylococcus aureus infections. J Clin Microbiol. 2007;45(1):127–133. doi: 10.1128/JCM.01866-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raspanti CG, Bonetto CC, Vissio C, Pellegrino MS, Reinoso EB, Dieser SA, et al. Prevalence and antibiotic susceptibility of coagulase-negative Staphylococcus species from bovine subclinical mastitis in dairy herds in the central region of Argentina. Rev Argent Microbiol. 2016;48(1):50–56. doi: 10.1016/j.ram.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 22.Li L, Zhou L, Wang L, Xue H, Zhao X. Characterization of methicillin-resistant and -susceptible staphylococcal isolates from bovine milk in northwestern china. PLoS One. 2015;10(3):e0116699. doi: 10.1371/journal.pone.0116699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nyman AK, Fasth C, Waller KP. Intramammary infections with different non-aureus staphylococci in dairy cows. J Dairy Sci. 2018;101(2):1403–1418. doi: 10.3168/jds.2017-13467. [DOI] [PubMed] [Google Scholar]
- 24.Lange CC, Brito MA, Reis DR, Machado MA, Guimarães AS, Azevedo AL, et al. Species-level identification of staphylococci isolated from bovine mastitis in Brazil using partial 16S rRNA sequencing. Vet Microbiol. 2015;176(3-4):382–388. doi: 10.1016/j.vetmic.2015.01.024. [DOI] [PubMed] [Google Scholar]
- 25.Srednik ME, Tremblay YD, Labrie J, Archambault M, Jacques M, Fernández Cirelli A, et al. Biofilm formation and antimicrobial resistance genes of coagulase-negative staphylococci isolated from cows with mastitis in Argentina. FEMS Microbiol Lett. 2017;364(8):fnx001. doi: 10.1093/femsle/fnx001. [DOI] [PubMed] [Google Scholar]
- 26.Felipe V, Morgante CA, Somale PS, Varroni F, Zingaretti ML, Bachetti RA, et al. Evaluation of the biofilm forming ability and its associated genes in Staphylococcus species isolates from bovine mastitis in Argentinean dairy farms. Microb Pathog. 2017;104:278–286. doi: 10.1016/j.micpath.2017.01.047. [DOI] [PubMed] [Google Scholar]
- 27.Jiménez Velásquez SD, Torres Higuera LD, Parra Arango JL, Rodríguez Bautista JL, García Castro FE, Patiño Burbano RE. Perfil de resistencia antimicrobiana en aislamientos de Staphylococcus spp. obtenidos de leche bovina en Colombia. Rev Argent Microbiol. 2020;52(2):121–130. doi: 10.1016/j.ram.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 28.Bhat AM, Soodan JS, Singh R, Dhobi IA, Hussain T, Dar MY, et al. Incidence of bovine clinical mastitis in Jammu region and antibiogram of isolated pathogens. Vet World. 2017;10(8):984–989. doi: 10.14202/vetworld.2017.984-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Odierno L, Calvinho L, Traverssa P, Lasagno M, Bogni C, Reinoso E. Conventional identification of Streptococcus uberis isolated from bovine mastitis in Argentinean dairy herds. J Dairy Sci. 2006;89(10):3886–3890. doi: 10.3168/jds.S0022-0302(06)72431-6. [DOI] [PubMed] [Google Scholar]
- 30.Bochniarz M, Wawron W. Antibiotic susceptibility of methicillin-resistant and methicillin-susceptible coagulase-negative staphylococci isolated from bovine mastitis. Pol J Vet Sci. 2011;14(3):405–410. doi: 10.2478/v10181-011-0060-5. [DOI] [PubMed] [Google Scholar]
- 31.Pu W, Su Y, Li J, Li C, Yang Z, Deng H, et al. High incidence of oxacillin-susceptible mecA-positive Staphylococcus aureus (OS-MRSA) associated with bovine mastitis in China. PLoS One. 2014;9(2):e88134. doi: 10.1371/journal.pone.0088134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soares L, Pereira I, Pribul B, Oliva M, Coelho S, Souza M. Antimicrobial resistance and detection of mecA and blaZ genes in coagulase-negative Staphylococcus isolated from bovine mastitis. Pesqui Vet Bras. 2012;32(8):692–696. [Google Scholar]
- 33.Dorneles EM, Fonseca MD, Abreu JA, Lage AP, Brito MA, Pereira CR, et al. Genetic diversity and antimicrobial resistance in Staphylococcus aureus and coagulase-negative Staphylococcus isolates from bovine mastitis in Minas Gerais, Brazil. MicrobiologyOpen. 2019;8(5):e00736. doi: 10.1002/mbo3.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Srednik ME, Usongo V, Lépine S, Janvier X, Archambault M, Gentilini ER. Characterisation of Staphylococcus aureus strains isolated from mastitis bovine milk in Argentina. J Dairy Res. 2018;85(1):57–63. doi: 10.1017/S0022029917000851. [DOI] [PubMed] [Google Scholar]
- 35.Srednik ME, Crespi E, Testorelli MF, Puigdevall T, Pereyra AM, Rumi MV, et al. First isolation of a methicillin-resistant Staphylococcus aureus from bovine mastitis in Argentina. Vet Anim Sci. 2018;7:100043. doi: 10.1016/j.vas.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Denamiel G, Carloni G, Llorente P, Gentilini E. Mastitis Bovina: prevalencia microbiana y perfil de resistencia en cocos gram positivos. Rev Med Vet. 2007;87(6):233–235. [Google Scholar]
- 37.Tyson GH, Li C, Ayers S, McDermott PF, Zhao S. Using whole-genome sequencing to determine appropriate streptomycin epidemiological cutoffs for Salmonella and Escherichia coli. FEMS Microbiol Lett. 2016;363(4):fnw009. doi: 10.1093/femsle/fnw009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cattoir V. In: Streptococcus pyogenes: Basic Biology to Clinical Manifestations. Ferretti JJ, Stevens DL, Fischetti VA, editors. Oklahoma City: University of Oklahoma Health Sciences Center; 2016. Mechanisms of antibiotic resistance. Available from: https://www.ncbi.nlm.nih.gov/books/NBK333414/ [PubMed] [Google Scholar]
- 39.Li T, Lu H, Wang X, Gao Q, Dai Y, Shang J, et al. Molecular Characteristics of Staphylococcus aureus Causing Bovine Mastitis between 2014 and 2015. Front Cell Infect Microbiol. 2017;7:127. doi: 10.3389/fcimb.2017.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Basanisi MG, La Bella G, Nobili G, Franconieri I, La Salandra G. Genotyping of methicillin-resistant Staphylococcus aureus (MRSA) isolated from milk and dairy products in South Italy. Food Microbiol. 2017;62:141–146. doi: 10.1016/j.fm.2016.10.020. [DOI] [PubMed] [Google Scholar]
- 41.Frey Y, Rodriguez JP, Thomann A, Schwendener S, Perreten V. Genetic characterization of antimicrobial resistance in coagulase-negative staphylococci from bovine mastitis milk. J Dairy Sci. 2013;96(4):2247–2257. doi: 10.3168/jds.2012-6091. [DOI] [PubMed] [Google Scholar]
- 42.Asadollahi P, Farahani NN, Mirzaii M, Khoramrooz SS, van Belkum A, Asadollahi K, et al. Distribution of the most prevalent Spa types among clinical isolates of methicillin-resistant and -susceptible Staphylococcus aureus around the world: a review. Front Microbiol. 2018;9:163. doi: 10.3389/fmicb.2018.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]