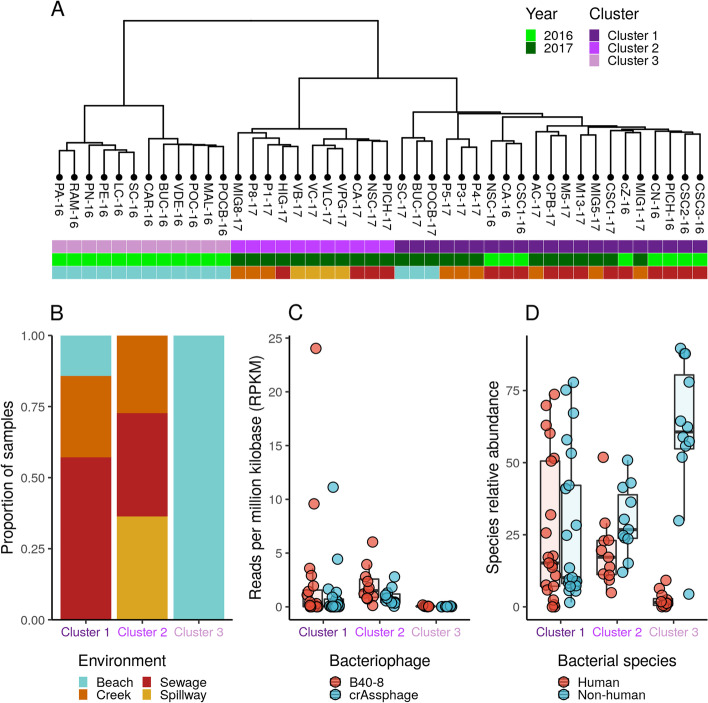
Fig. 1.
Dissemination of human bacteria in different niches of urban water environments. A Clustering analysis based on k-mers frequency from urban water metagenomes (n = 44). Samples are colored according to their belonging to each main cluster (clusters 1, 2, and 3), sampling year, and environment type according to their collection site (beach, sewage, creek, or spillway). B Barplot showing the proportion of samples belonging to each metagenomic cluster stratified by type of environment. C Metagenomic quantification (measured as reads per kilobase-million, RPKM) of human-gut-microbiota-associated bacteriophages B40-8 and crAssphage in samples belonging to each cluster. D Metagenomic quantification (measured as relative abundances) of bacterial species that are associated or not associated to the human gut microbiota in samples belonging to each cluster