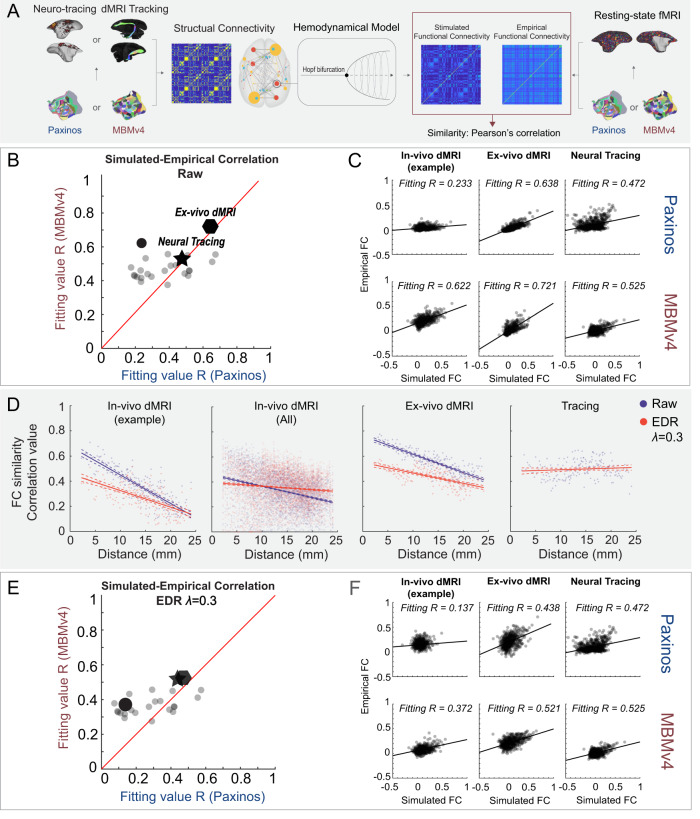
Fig. 7. A computational framework links the structural-functional connectivity according to different parcellations.
A Application of whole-brain modeling, including an estimation of structural connectivity from neuronal tracing, diffusion MRI (in vivo or ex vivo) according to the Paxinos atlas or MBMv4, simulation of functional connectivity from structural connectivity by the Hopf bifurcation hemodynamic functions, and a similarity measure with empirical connectivity from resting-state fMRI. B Model fitting comparison in different spatial scales using the Paxinos versus the MBMv4 atlases. Individual examples using in vivo diffusion MRI (round dots; an example animal marked in black dot), ex vivo diffusion MRI (polygon), and neuronal tracing (star) show higher fitting values for MBMv4 versus the Paxinos atlas. C Examples of correlations between the simulated and empirical functional connectivity from B; solid black lines represent marginal regression lines. D Model fitting comparisons with (red) and without (blue) an exponential distance rule (EDR) correction. From left to right, the plots present the simulation results obtained with in vivo diffusion MRI from the example animal (the linear regression values R = 0.689, p = 9.27e-50 for the blue line; R = 0.413, p = 1.28e-23 for the red); in vivo diffusion MRI from all animals (R = 0.11, p = 6.05e-114 for the blue; R = 0.011, p = 4.17e-13 for the red); the ex vivo diffusion MRI (R = 0.7, p = 2.69e-51 for the blue; R = 0.395, p = 2.19e-22 for the red); and the neuronal-tracing dataset (R = 0.009, p = 0.02). The line plots are presented as mean values ± 95% C.I. Dashed lines represent the 95% confidence interval. E Model fitting comparison with EDR correction between the Paxinos atlas and MBMv4 in different spatial scales. F Examples of correlation between the simulated and empirical functional connectivity from E.