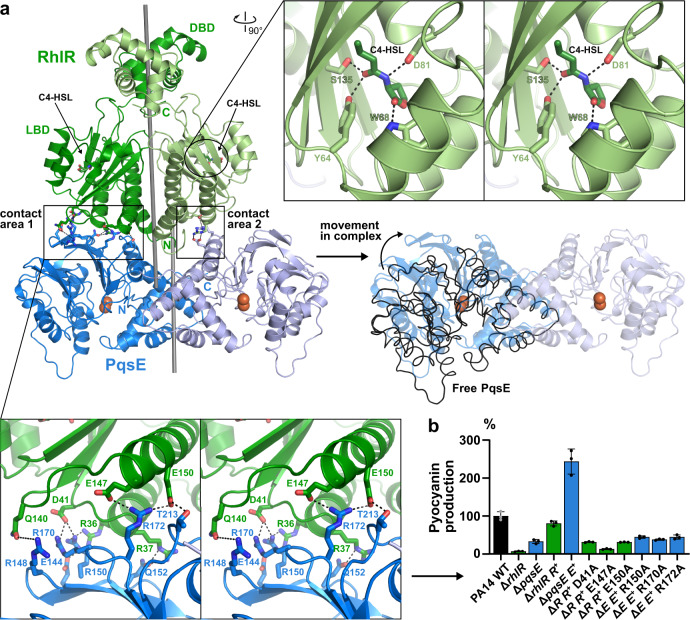
Fig. 3. Structure of the PqsE/RhlR complex.
a Crystal structure of the PqsE/RhlR:C4-HSL complex. DBD and LBD denote the DNA- and ligand-binding domains of RhlR, the two-fold symmetry axes of RhlR and PqsE are shown as sticks (calculated with draw_rotation_axis.py in PyMOL73). Red spheres represent two iron atoms bound to the active center of PqsE. The magnified area at the top right shows a cross-eyed stereo plot of polar interactions between C4-HSL and RhlR, the magnified area below demonstrates polar interactions between RhlR and PqsE in the larger contact area 1. Complex formation with RhlR distorts the PqsE dimer with respect to the free enzyme (black lines, middle-right panel), leading to movements of more than 10 Å at extreme positions. b Mutation of selected residues in contact area 1 impairs the production of pyocyanin in pUCP24-supplemented (indicated by R+ or E+, respectively) ΔrhlR and ΔpqsE mutants of P. aeruginosa PA14. Mean values with error bars indicating the standard deviation of three independent measurements are shown. Source data are provided as a Source Data file.