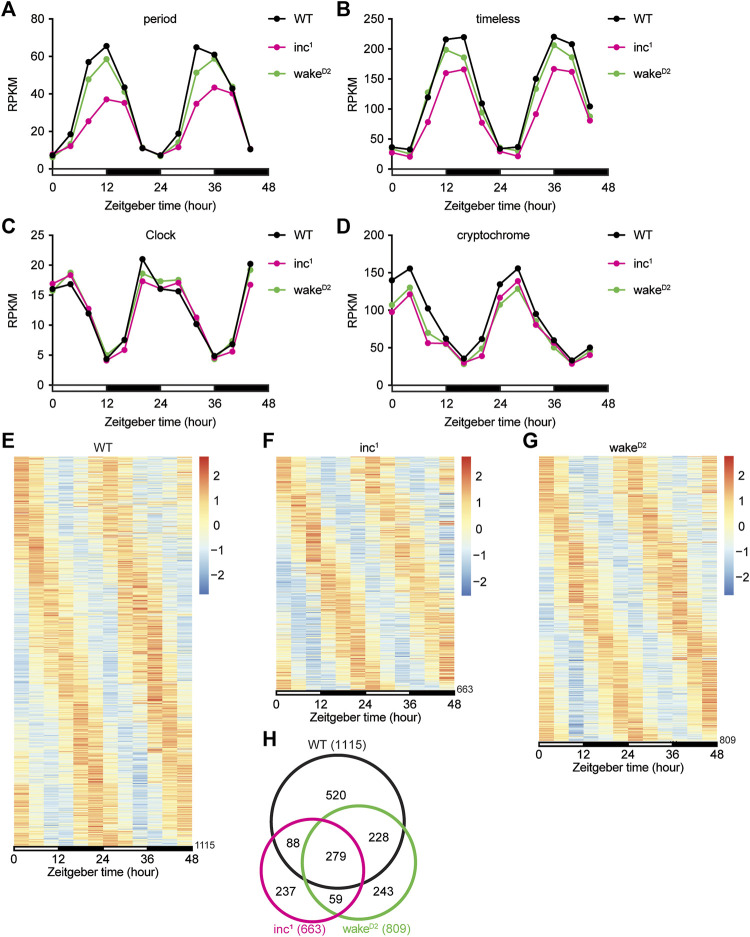
FIGURE 2.
Chronic sleep loss by inc or wake mutations disrupts circadian gene oscillation. (A–D) Reads Per Kilobase of transcript, per Million mapped reads (RPKM) of period (A), timeless (B), Clock (C), and cryptochrome (D) for WT (black), inc 1 (magenta), and wake D2 (green). x axis represents two 12-hour light/12-hour dark (LD) cycles with white bars representing daytime and black bars representing nighttime. For period, **** inc 1 and n.s. wake D2 . For timeless, **** inc 1 and n.s. wake D2 . For Clock, n.s. inc 1 and wake D2 . For cryptochrome, **** inc 1 and wake D2 . All comparisons are against WT, using Benjamini-Hochberg adjusted p values calculated by DESeq2 for time-series analysis. ****p < 0.0001; n.s., not significant. (E–G) Heatmaps of rhythmic genes in WT (E), inc 1 (F), wake D2 (G) groups. Each row represents one gene, and each column represents one timepoint. Expression levels are normalized to each gene itself and data are aligned according to the peak expression time calculated by JTK cycle. The blocked horizontal bars below the panel correspond to the light-dark cycle. (H) Intersection of rhythmic genes in WT (black), inc 1 (magenta), and wake D2 (green).